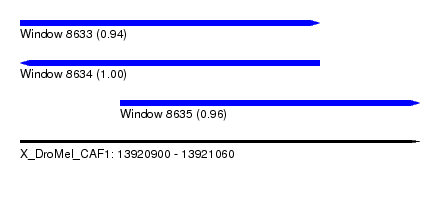
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,920,900 – 13,921,060 |
| Length | 160 |
| Max. P | 0.999879 |

| Location | 13,920,900 – 13,921,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -39.54 |
| Energy contribution | -39.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13920900 120 + 22224390 AAUGACAUGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCCAAGCAAUCUGAGCCACGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUU .((((..(((.((....)).))).....(.((((.((((((((((...........((....))(((((........)))))........))))))).)))...)))))....))))... ( -39.60) >DroSec_CAF1 1154 120 + 1 AAUGACACGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCCAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUU .((((((.(((.....((((......)))).))).))((((.........(((((((((.((..(((((........)))))....))..)))))))))......))))....))))... ( -40.56) >DroSim_CAF1 3139 120 + 1 AAUGACAUGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCCAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUU .((((((.(((.....((((......)))).))).))((((.........(((((((((.((..(((((........)))))....))..)))))))))......))))....))))... ( -40.56) >DroEre_CAF1 2651 119 + 1 AAUGACAUGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCC-AGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUU .((((((.(((.....((((......)))).))).))((((-........(((((((((.((..(((((........)))))....))..)))))))))......))))....))))... ( -41.14) >DroYak_CAF1 2585 120 + 1 AAUGACAUGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCCAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUA .((((((.(((.....((((......)))).))).))((((.........(((((((((.((..(((((........)))))....))..)))))))))......))))....))))... ( -40.56) >consensus AAUGACAUGGGAGGAAGCUACCAACUUAGCGCCCUUGGGCCAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUU .((((((.(((.....((((......)))).))).))((((.........(((((((((.((..(((((........)))))....))..)))))))))......))))....))))... (-39.54 = -39.74 + 0.20)


| Location | 13,920,900 – 13,921,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -49.38 |
| Energy contribution | -49.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13920900 120 - 22224390 AAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACGUGGCUCAGAUUGCUUGGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCAUGUCAUU ..((((((((.(((((...(((.(((((((((.......((((..(.((((((....)))))))..))))...)))))))))))).))))((((......))))......).)))))))) ( -52.90) >DroSec_CAF1 1154 120 - 1 AAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACUUGGCUCAGAUUGCUUGGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCGUGUCAUU ..(((((..(.(((((...(((.(((((((((.......((((((.(((((((....)))))))))))))...)))))))))))).))))((((......))))......).)..))))) ( -50.90) >DroSim_CAF1 3139 120 - 1 AAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACUUGGCUCAGAUUGCUUGGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCAUGUCAUU ..((((((((.(((((...(((.(((((((((.......((((((.(((((((....)))))))))))))...)))))))))))).))))((((......))))......).)))))))) ( -51.30) >DroEre_CAF1 2651 119 - 1 AAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACUUGGCUCAGAUUGCU-GGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCAUGUCAUU ..((((((((.(((((...(((.((((.((((.......((((((.(((((((....)))))))))))))...)))))-)))))).))))((((......))))......).)))))))) ( -46.60) >DroYak_CAF1 2585 120 - 1 UAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACUUGGCUCAGAUUGCUUGGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCAUGUCAUU ..((((((((.(((((...(((.(((((((((.......((((((.(((((((....)))))))))))))...)))))))))))).))))((((......))))......).)))))))) ( -51.30) >consensus AAGAUGAUGUUGGCCCGAAUGGAGCCAAGCGAGAAAAUGGAGCUGCAGUUGGCCCCCGUCAACUUGGCUCAGAUUGCUUGGCCCAAGGGCGCUAAGUUGGUAGCUUCCUCCCAUGUCAUU ..(((((..(.(((((...(((.(((((((((.......((((((.(((((((....)))))))))))))...)))))))))))).))))((((......))))......).)..))))) (-49.38 = -49.62 + 0.24)

| Location | 13,920,940 – 13,921,060 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -38.59 |
| Consensus MFE | -34.13 |
| Energy contribution | -34.57 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13920940 120 + 22224390 CAAGCAAUCUGAGCCACGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUUCUUCACUUCGUCUUCGCGUUUUGGCCUCAAGUCGGUUGGC (((.(.(..((((((((((.(((((((((........)))))........(.(((((((.....))))))))................))))...))))...)).))))..).).))).. ( -34.40) >DroSec_CAF1 1194 120 + 1 CAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUUCUUCUCUUCGCCUUCGCGUUUUGGCCUCAAGUCGGUUGGC ....(((((.(((((((((.((..(((((........)))))....))..))))))))).....(((((((.................(((....)))..)))))))......))))).. ( -36.81) >DroSim_CAF1 3179 120 + 1 CAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUUCUUCUCUUCGCCUUCGCGUUUUGGCCUCAAGUCGGUUGGC ....(((((.(((((((((.((..(((((........)))))....))..))))))))).....(((((((.................(((....)))..)))))))......))))).. ( -36.81) >DroEre_CAF1 2691 119 + 1 C-AGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUUCUUCUCUUCGCCUUCGCGUUUUGGCCUGAAGUCGGUUGCG .-.((((((.(((((((((.((..(((((........)))))....))..)))))))))..((((((((((.................(((....)))..))))))))))...)))))). ( -46.01) >DroYak_CAF1 2625 120 + 1 CAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUACUUCUCUUAGCCUUCGCGUUUUGGCCUCAAGUCGCUUGGC (((((.(..(((((((((.((.(((((((.......(((...........)))((((((.....))))))...................))))))))).))))).))))..).))))).. ( -38.90) >consensus CAAGCAAUCUGAGCCAAGUUGACGGGGGCCAACUGCAGCUCCAUUUUCUCGCUUGGCUCCAUUCGGGCCAACAUCAUCUUCUUCUCUUCGCCUUCGCGUUUUGGCCUCAAGUCGGUUGGC ....(((((.(((((((((.((..(((((........)))))....))..))))))))).....(((((((.................(((....)))..)))))))......))))).. (-34.13 = -34.57 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:10 2006