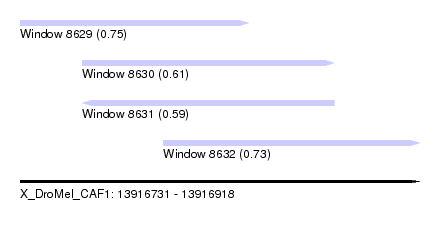
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,916,731 – 13,916,918 |
| Length | 187 |
| Max. P | 0.754195 |

| Location | 13,916,731 – 13,916,838 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -19.15 |
| Energy contribution | -20.40 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13916731 107 + 22224390 AAUCCAAGAGCUAGCUAACUGCAGCUCAAGAGCGCUGCCAAAGAGAGCGCCACUUAG--GCACUGAUUGAGAGAGCAAUCUGGCACGAUGCGAGAGCGGGAGCUUAAAA ..(((..(((((.((.....))))))).....((.(((((.....((.(((.....)--)).))(((((......)))))))))))).(((....))))))........ ( -35.10) >DroSec_CAF1 17055 107 + 1 AAUCCAAGAGCUCCCAAACUGCAGCUCAAGAGCGCUGCAAAAGAGAGCGCGACUUAG--GCACUGAUUGGGAGAACAAUCUGGCACGUUGCGAGAGCGGGAGCUUAAAA .......((((((((.((((((.((..(((.(((((.(......))))))..)))..--)).(.(((((......))))).)))).)))((....)))))))))).... ( -42.70) >DroEre_CAF1 12948 105 + 1 AAUCCAAGAGCUCGCUCGCUGCAGCUCAAGAGCGCUGGAAAAGAGAGCCCGACUCAG--AGACGGAUUGAGAGUAG-AACUAGCACGUUGCGAGCGCGAGAGUU-UAAA .......((((((((((((.((.(((....)))(((((....(......)..(((((--.......))))).....-..)))))..)).))))))....)))))-)... ( -29.60) >DroYak_CAF1 27740 109 + 1 AAUCCGAGAGCUCGCAAGCUGCAGCUCAAGAGCGCUGAAAAAGAGAGCUCAACUCAGGCACACGGAUUGCGAGUGCAAACUGCUACGUUGCGAGCAUAAGAGCUUCAAA .........(((((((((..((((.....((((.((.....))...)))).......((((.((.....)).))))...))))..).)))))))).............. ( -34.90) >consensus AAUCCAAGAGCUCGCAAACUGCAGCUCAAGAGCGCUGCAAAAGAGAGCGCGACUCAG__ACACGGAUUGAGAGAACAAACUGGCACGUUGCGAGAGCGAGAGCUUAAAA .......((((((....((((((((......(((((.........))))).................((..(((....)))..)).))))).)))....)))))).... (-19.15 = -20.40 + 1.25)

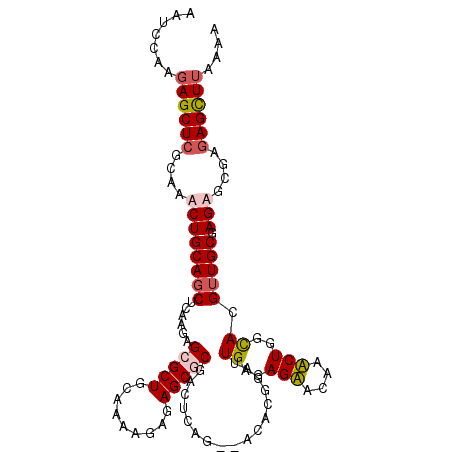

| Location | 13,916,760 – 13,916,878 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13916760 118 + 22224390 GAGCGCUGCCAAAGAGAGCGCCACUUAG--GCACUGAUUGAGAGAGCAAUCUGGCACGAUGCGAGAGCGGGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGC ..((((((((..(((..(((((.....)--)).....((....))))..))).((.((((((..(((..(((((((............))))))).)))..))))))..)).)))))))) ( -46.10) >DroSec_CAF1 17084 118 + 1 GAGCGCUGCAAAAGAGAGCGCGACUUAG--GCACUGAUUGGGAGAACAAUCUGGCACGUUGCGAGAGCGGGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGC ..(((((((......(((.(.(((((..--((...(((((......)))))..)).(((((((.((((....))))....))))).))))))).).)))..((......))..))))))) ( -42.90) >DroEre_CAF1 12977 116 + 1 GAGCGCUGGAAAAGAGAGCCCGACUCAG--AGACGGAUUGAGAGUAG-AACUAGCACGUUGCGAGCGCGAGAGUU-UAAAUGCAAUCGCAGUCUCACUCAAGCGUCGCAGCGGGCAGCGC ..((((((.....(..((....((((.(--(......))..))))..-..))..).((((((((.(((..((((.-....(((....))).....))))..)))))))))))..)))))) ( -41.60) >DroYak_CAF1 27769 120 + 1 GAGCGCUGAAAAAGAGAGCUCAACUCAGGCACACGGAUUGCGAGUGCAAACUGCUACGUUGCGAGCAUAAGAGCUUCAAAUGCAAUCGAAGUUUCACUCAGGCGUCGCAGCAGGCAGCGC ((((.((.....))...)))).......((((.((.....)).))))...(((((..(((((((((....(((((((..........))))))).......)).))))))).)))))... ( -38.00) >consensus GAGCGCUGCAAAAGAGAGCGCGACUCAG__ACACGGAUUGAGAGAACAAACUGGCACGUUGCGAGAGCGAGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGC ..(((((((......(((.....)))............((..(((....)))..)).(((((((...(((..((.......))..)))..((((.....)))).)))))))..))))))) (-25.31 = -25.25 + -0.06)


| Location | 13,916,760 – 13,916,878 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -41.13 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13916760 118 - 22224390 GCGCUGCCUGCUGCGACGCUUGAGUGAAACUUCGAUUGCAUUUUAAGCUCCCGCUCUCGCAUCGUGCCAGAUUGCUCUCUCAAUCAGUGC--CUAAGUGGCGCUCUCUUUGGCAGCGCUC ((((((((.((.((((.((..(((((...(((.((......)).)))....)))))..)).))))))..(((((......)))))(((((--(.....))))))......)))))))).. ( -44.90) >DroSec_CAF1 17084 118 - 1 GCGCUGCCUGCUGCGACGCUUGAGUGAAACUUCGAUUGCAUUUUAAGCUCCCGCUCUCGCAACGUGCCAGAUUGUUCUCCCAAUCAGUGC--CUAAGUCGCGCUCUCUUUUGCAGCGCUC (((((((..((.(((((((..(((((...(((.((......)).)))....)))))..))...(..(..(((((......))))).)..)--....)))))))........))))))).. ( -37.80) >DroEre_CAF1 12977 116 - 1 GCGCUGCCCGCUGCGACGCUUGAGUGAGACUGCGAUUGCAUUUA-AACUCUCGCGCUCGCAACGUGCUAGUU-CUACUCUCAAUCCGUCU--CUGAGUCGGGCUCUCUUUUCCAGCGCUC (((((((.((.((((((((..((((..((.(((....))).)).-.))))..))).))))).)).)).((((-(.((((...........--..)))).))))).........))))).. ( -40.62) >DroYak_CAF1 27769 120 - 1 GCGCUGCCUGCUGCGACGCCUGAGUGAAACUUCGAUUGCAUUUGAAGCUCUUAUGCUCGCAACGUAGCAGUUUGCACUCGCAAUCCGUGUGCCUGAGUUGAGCUCUCUUUUUCAGCGCUC ((((((.((((((((..((..(((((.((((((((......))))))...)).)))))))..))))))))...((((.((.....)).))))..(((.....))).......)))))).. ( -41.20) >consensus GCGCUGCCUGCUGCGACGCUUGAGUGAAACUUCGAUUGCAUUUUAAGCUCCCGCGCUCGCAACGUGCCAGAUUGCACUCUCAAUCAGUGC__CUAAGUCGCGCUCUCUUUUGCAGCGCUC (((((((.((.((((((((..((((.....................))))..))).))))).))......................((((.........))))........))))))).. (-23.36 = -23.80 + 0.44)

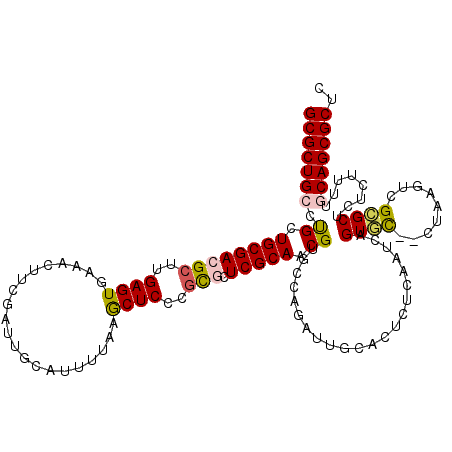
| Location | 13,916,798 – 13,916,918 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -27.01 |
| Energy contribution | -31.20 |
| Covariance contribution | 4.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13916798 120 + 22224390 AGAGAGCAAUCUGGCACGAUGCGAGAGCGGGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGCAGUCGGCGUUGCAGCUCAAAGAGAACUGUGUCGCAAAAGA .(((.((......)).(((((((.((((....))))....))).))))........)))..(((.(((((...(((((((.....)))))))..(((...)))..))))).)))...... ( -40.30) >DroSec_CAF1 17122 120 + 1 GGAGAACAAUCUGGCACGUUGCGAGAGCGGGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGCAGCCGGCGUUGCAGCUGAAAGAGAACUGUGACGCGAAAGA .(((..(..(((.((.....)).)))..)(((((((............))))))).)))..(((((((((((((((((((.....)))))))..)))........)))))))))...... ( -40.80) >DroEre_CAF1 13015 118 + 1 AGAGUAG-AACUAGCACGUUGCGAGCGCGAGAGUU-UAAAUGCAAUCGCAGUCUCACUCAAGCGUCGCAGCGGGCAGCGCAGCCGGCGUUGCAGCUCGAAGAGAACAGAUUCGCGAAAGA .((((..-........((((((((.(((..((((.-....(((....))).....))))..))))))))))).(((((((.....))))))).))))................(....). ( -44.40) >DroYak_CAF1 27809 120 + 1 CGAGUGCAAACUGCUACGUUGCGAGCAUAAGAGCUUCAAAUGCAAUCGAAGUUUCACUCAGGCGUCGCAGCAGGCAGCGCAGCCGGCGUUGCAGCUCAAAGAGAACUGUUUCGCCAAAGA .(((((.(((((....((((((((((......)))).....)))).)).)))))))))).((((..((((...(((((((.....)))))))..(((...)))..))))..))))..... ( -43.10) >consensus AGAGAACAAACUGGCACGUUGCGAGAGCGAGAGCUUAAAAUGCAAUCGAAGUUUCACUCAAGCGUCGCAGCAGGCAGCGCAGCCGGCGUUGCAGCUCAAAGAGAACUGUGUCGCGAAAGA .(((((.(((((.....((((((.((((....))))....))))))...))))))))))..(((.(((((...(((((((.....)))))))..(((...)))..))))).)))...... (-27.01 = -31.20 + 4.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:07 2006