| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,525,635 – 1,525,742 |
| Length | 107 |
| Max. P | 0.524727 |

| Location | 1,525,635 – 1,525,742 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.51 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
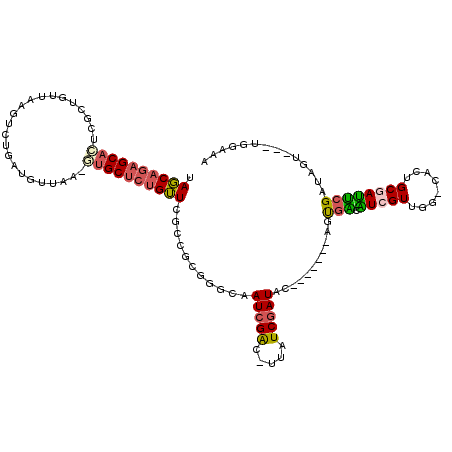
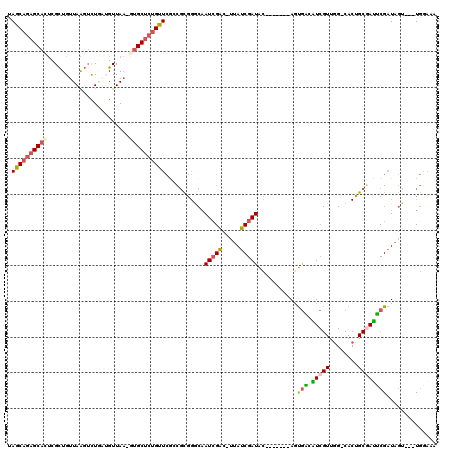
>X_DroMel_CAF1 1525635 107 - 22224390 CAGCAGAGCACUCGCUGUUAAGUCUGAUGUUAA-AUGCUCUGUUCGCCGCGGGCAAUCGAC-UUAUCGAUAC-------AGUGACAUCGUUUG-CACUGCGAUUCGAUAGU---UGGCAA .(((((((((...((.((((....))))))...-.)))))))))....((.(((.(((((.-..((((...(-------((((.((.....))-)))))))))))))).))---).)).. ( -32.60) >DroPse_CAF1 4157 91 - 1 AAUC---GCAGCUGUUGUUAGUUUUGGUGUUACACAGCACUGAUUCCUU-----CAUCGAUAUUAUCGAUGC-------AACAA-GU-------------GAUUCGCUACUGAAUCGAAU ..((---...(((((((...(..(..(((((....)))))..)..)...-----(((((((...))))))))-------))).)-))-------------((((((....)))))))).. ( -22.10) >DroSec_CAF1 15059 107 - 1 UAGCAGAGCACUCGCUGUUAAGUCUGAUGUUAA-GUGCUCUGUUCGGCGCGGGCAAUCGAC-UUAUCGAUAC-------AAUGAAGUCGUUGG-CACUGCGAUUCGAUAGU---UGGAGA ..((((((((((.(..((((....))))..).)-)))))))))(((.(((((((.(((((.-...))))).(-------((((....))))))-).)))))...)))....---...... ( -36.10) >DroSim_CAF1 16334 107 - 1 UAGCAGAGCACUUGCUGUUAAGUCUGAUGUUAA-GUGCUCUGUUCGGCGCGGGCAAUCGAC-UUAUCGAUAC-------AGUGAAGUCGUUGG-CACUGCGACUCGAUAGU---UGGAGA .(((((((((((((..((((....))))..)))-))))))))))...(((((((...((((-((..(.....-------.)..))))))...)-).))))).(((.(....---).))). ( -40.80) >DroEre_CAF1 15138 112 - 1 UAGCAGAGCAGUCGCUGUUAAGUCU---GUUAA-GUGCUCUGUUCGCCGUGGACAAUCGGC-UCAUCGAUACGCAUAACAGUGCCAUCGUUGGACACUGCGAUGCGAUAGU---UGUAAG ((((.((((((.((((.........---....)-)))..))))))((((........))))-.........(((((..((((((((....))).)))))..)))))...))---)).... ( -37.32) >DroYak_CAF1 16666 112 - 1 UAGCAGAGCACUCGCUGUUAGGACU---GUUAA-GUGCUCUGUUCACCGCGGGCAAUAGAC-UUAUCGAUACGCGCAAUAGUGUCAUCGUCGGACACCGCUAUACGAUAGU---UGGAAA .(((((((((((.((.((....)).---))..)-))))))))))..(((....)....(((-(.(((((((.(((.....(((((.......))))))))))).)))))))---)))... ( -32.40) >consensus UAGCAGAGCACUCGCUGUUAAGUCUGAUGUUAA_GUGCUCUGUUCGCCGCGGGCAAUCGAC_UUAUCGAUAC_______AGUGACAUCGUUGG_CACUGCGAUUCGAUAGU___UGGAAA .((((((((((.......................))))))))))...........(((((.....)))))...........(((.(((((........)))))))).............. (-14.94 = -15.08 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:06 2006