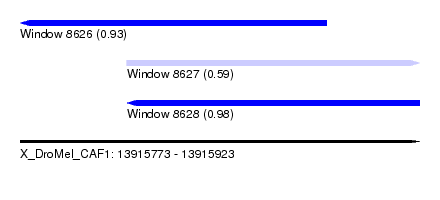
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,915,773 – 13,915,923 |
| Length | 150 |
| Max. P | 0.982159 |

| Location | 13,915,773 – 13,915,888 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -22.46 |
| Energy contribution | -21.86 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
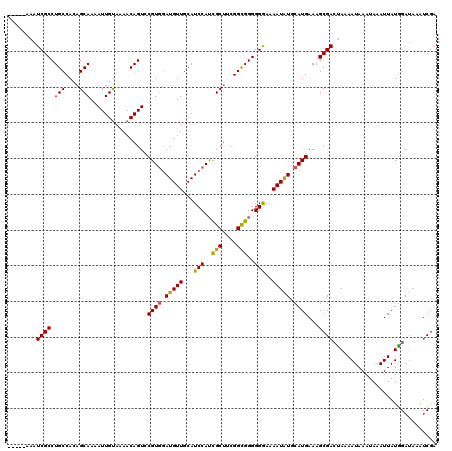
>X_DroMel_CAF1 13915773 115 - 22224390 -----AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUCCGUGGAUGUUGCAUCCAUUGCUGCGGCGGGGGGAAAAUAUGCAUGAAAGCGAAUAAAAUAAAUAAAUUAUGGAAAAAUCGA -----...((.((((((.((((((.((((.....))))...((((((...)))))))))))).)))))).))......(((......))).............................. ( -31.70) >DroSec_CAF1 16085 115 - 1 -----AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUUCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUCCAUGAAAGCGACUAAAAUAAAUAAAUUAUAGAUAAAUCGA -----...((((.(((....)))..............((((((((((((...(((.((((....)))))))...)))))))))))).))))(((.(((......))).)))......... ( -33.80) >DroSim_CAF1 38053 113 - 1 -----AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUUCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUCCAUGAAAGCGACUAAAAUAAAUAAAUUAUGGAU--AUCGA -----...((((.(((....)))..............((((((((((((...(((.((((....)))))))...)))))))))))).))))(((.(((......))).)))..--..... ( -33.50) >DroEre_CAF1 12000 120 - 1 UCGAAAAAUCGCCUGCCAUCGCAAAAUUGUACAACAGUCCGUAAAUGUUGCAUCCAUCGCUUCGGUGGGGGAAAAAUGUGCAUGAAAGCGACCAAAAUAAAUAAAUUAUGGAUAAAUCGA ((((....((((.(((....)))....((((((....((((((.....))).(((((((...))))))))))....)))))).....))))(((.(((......))).))).....)))) ( -28.90) >DroYak_CAF1 26743 114 - 1 -----AAAUCGCCUGCCACAGCAAAAUUGCACAACAGUCCGUGGAUGUUGCAUCCAGCGCUUCGGUGGGGGAA-AAUAUGCAUGAAAGCGACUAAAAUAAAUAAAUUAUGGAUAAAUCGA -----...(((((((.....(((....)))....)))..((((.(((((...(((..(((....)))..))).-))))).))))...))))(((.(((......))).)))......... ( -24.90) >consensus _____AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUCCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUGCAUGAAAGCGACUAAAAUAAAUAAAUUAUGGAUAAAUCGA ........((((.(((....)))................((((.(((((...(((..(((....)))..)))..))))).))))...))))............................. (-22.46 = -21.86 + -0.60)

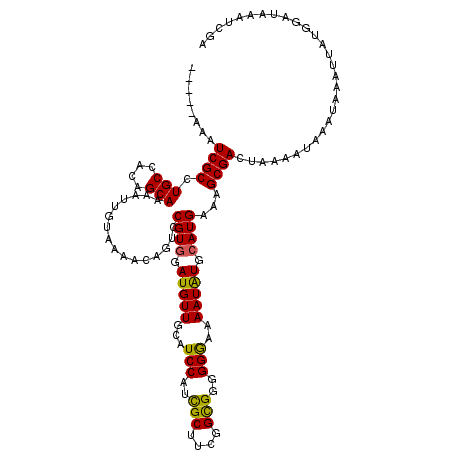
| Location | 13,915,813 – 13,915,923 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.37 |
| Mean single sequence MFE | -36.89 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13915813 110 + 22224390 CAUAUUUUCCCCCCGCCGCAGCAAUGGAUGCAACAUCCACGGACUGUUUUACAAUUUUGCUGUGGCAGGCGAUUU-------U-CCGAUU-UUCCACCGGCCUUCC-AUUCGCUGGGGGA .........(((((((((((((((((((((...)))))).....((.....))...)))))))))).(((((...-------.-(((...-......)))......-..)))))))))). ( -41.90) >DroSec_CAF1 16125 110 + 1 GAUAUUUUCCCCCCGCCGAAGCGAUGGAUGCAACAUCCACGAACUGUUUUACAAUUUUGCUGUGGCAGGCGAUUU-------U-CCGAUUUUUCCACCGGCCUUCC-AUCCGCCGGGG-C ..........((((((((.(((((((((((...)))))).....((.....))...))))).)))).((((....-------.-..((....))....((....))-...))))))))-. ( -31.70) >DroSim_CAF1 38091 109 + 1 GAUAUUUUCCCCCCGCCGAAGCGAUGGAUGCAACAUCCACGAACUGUUUUACAAUUUUGCUGUGGCAGGCGAUUU-------U-CCGAUU-UUCCACCGGCCUUCC-AUCCGGCGGGG-C ..........((((((((.(((((((((((...)))))).....((.....))...)))))((((.((((.....-------.-......-........)))).))-)).))))))))-. ( -36.35) >DroEre_CAF1 12040 117 + 1 CACAUUUUUCCCCCACCGAAGCGAUGGAUGCAACAUUUACGGACUGUUGUACAAUUUUGCGAUGGCAGGCGAUUUUUCGAUUUUUCGAUU-UUCCACCGGCCUUCC-AUCCGCCGGGG-G .........(((((..((..((((..(.(((((((.........))))))))....))))(((((.((((((....((((....))))..-.)).....)))).))-)))))..))))-) ( -36.30) >DroYak_CAF1 26783 110 + 1 CAUAUU-UUCCCCCACCGAAGCGCUGGAUGCAACAUCCACGGACUGUUGUGCAAUUUUGCUGUGGCAGGCGAUUU-------UGUCGAUU-UUUCACCGGCCUUUCAAUUCGCUGGGG-G ......-..((((((.((((..(((((..((((((((....)).))))))((((..(((((......)))))..)-------))).....-.....))))).......)))).)))))-) ( -38.20) >consensus CAUAUUUUCCCCCCGCCGAAGCGAUGGAUGCAACAUCCACGGACUGUUUUACAAUUUUGCUGUGGCAGGCGAUUU_______U_CCGAUU_UUCCACCGGCCUUCC_AUCCGCCGGGG_C ..........((((((((.(((((((((((...)))))).....((.....))...))))).)))).((((.............(((..........)))..........)))))))).. (-23.70 = -23.74 + 0.04)

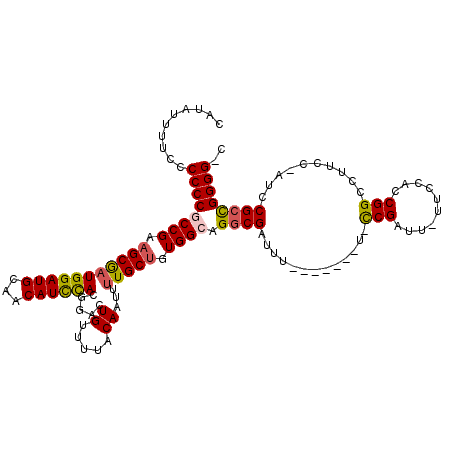
| Location | 13,915,813 – 13,915,923 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.37 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
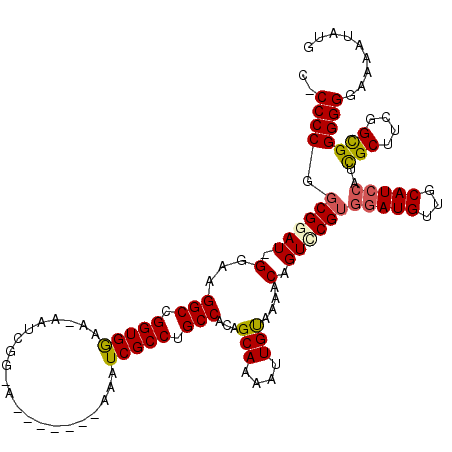
>X_DroMel_CAF1 13915813 110 - 22224390 UCCCCCAGCGAAU-GGAAGGCCGGUGGAA-AAUCGG-A-------AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUCCGUGGAUGUUGCAUCCAUUGCUGCGGCGGGGGGAAAAUAUG ((((((.((((..-......(((((....-.)))))-.-------...)))).((((.((((((.((((.....))))...((((((...)))))))))))).))))))))))....... ( -43.70) >DroSec_CAF1 16125 110 - 1 G-CCCCGGCGGAU-GGAAGGCCGGUGGAAAAAUCGG-A-------AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUUCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUC .-(((((((((.(-((....(((((......)))))-.-------...))).))))).......(((((.....))))).(((((((...)))))))(((....)))))))......... ( -38.60) >DroSim_CAF1 38091 109 - 1 G-CCCCGCCGGAU-GGAAGGCCGGUGGAA-AAUCGG-A-------AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUUCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUC .-(((((((((((-((.((((((((....-.))))(-.-------....))))).)))..((..(((((.....))))).(((((((...))))))).)))))))))))).......... ( -42.10) >DroEre_CAF1 12040 117 - 1 C-CCCCGGCGGAU-GGAAGGCCGGUGGAA-AAUCGAAAAAUCGAAAAAUCGCCUGCCAUCGCAAAAUUGUACAACAGUCCGUAAAUGUUGCAUCCAUCGCUUCGGUGGGGGAAAAAUGUG (-(((((.(((((-(((.(((.(((((..-..((((....))))....))))).)))...((((.....(((........)))....)))).))))))....)).))))))......... ( -43.60) >DroYak_CAF1 26783 110 - 1 C-CCCCAGCGAAUUGAAAGGCCGGUGAAA-AAUCGACA-------AAAUCGCCUGCCACAGCAAAAUUGCACAACAGUCCGUGGAUGUUGCAUCCAGCGCUUCGGUGGGGGAA-AAUAUG (-(((((.(((((((...(((.(((((..-........-------...))))).)))...(((....)))....)))).((((((((...)))))).))..))).))))))..-...... ( -39.32) >consensus C_CCCCGGCGGAU_GGAAGGCCGGUGGAA_AAUCGG_A_______AAAUCGCCUGCCACAGCAAAAUUGUAAAACAGUCCGUGGAUGUUGCAUCCAUCGCUUCGGCGGGGGGAAAAUAUG ..((((.((((((.(...(((.(((((.....................))))).)))...(((....)))....).))))))(((((...)))))..(((....)))))))......... (-30.08 = -30.28 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:04 2006