
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,914,523 – 13,914,664 |
| Length | 141 |
| Max. P | 0.992027 |

| Location | 13,914,523 – 13,914,625 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.48 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13914523 102 + 22224390 -------------UUUCGUUUUC-----AGGGUUCCCUGCGUGCCGCUGUGCUGCAGCUUUGGCUUAGUAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC -------------....(((((.-----.((((....(((((.....(((((((((((..((((........))))...)))))))))))......))))).......))))..))))). ( -39.60) >DroSec_CAF1 14768 102 + 1 -------------UUUCGUUUUC-----AGGGUUCCCUGCGUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC -------------....(((((.-----.((((....(((((.....(((((((((((..((((........))))...)))))))))))......))))).......))))..))))). ( -39.60) >DroSim_CAF1 36770 102 + 1 -------------UUUCGUUUUC-----AGGGUUCCCUGCGUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC -------------....(((((.-----.((((....(((((.....(((((((((((..((((........))))...)))))))))))......))))).......))))..))))). ( -39.60) >DroEre_CAF1 10625 102 + 1 -------------UUUCGUUUUC-----AGGGUUCCCUGCGUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC -------------....(((((.-----.((((....(((((.....(((((((((((..((((........))))...)))))))))))......))))).......))))..))))). ( -39.60) >DroWil_CAF1 46393 119 + 1 UUUCUGAUUCGGUUUUUGUUUUCUUGGCAGAGCU-GCUGCGUGCGGUUCUGUGGCAGUCUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCAGCAGCAGCA ..............((((((.....))))))(((-((((((..(((...((((..(((.((((((..((((((.....))))))))).))))))...))))...)))..).)))))))). ( -47.80) >DroYak_CAF1 25357 102 + 1 -------------UUUCGUUUUC-----AGGGUUCCCUGCGUGCCGCAGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC -------------....(((((.-----.((((....(((((......((((((((((..((((........))))...)))))))))).......))))).......))))..))))). ( -38.02) >consensus _____________UUUCGUUUUC_____AGGGUUCCCUGCGUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCC .............................((((....(((((.....(((((((((((..((((........))))...)))))))))))......))))).......))))........ (-33.06 = -33.48 + 0.42)



| Location | 13,914,545 – 13,914,664 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -36.50 |
| Energy contribution | -37.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13914545 119 + 22224390 GUGCCGCUGUGCUGCAGCUUUGGCUUAGUAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACCAAAGGCACCCAGCCACC-ACAUCAUGCC ((((.(((((((((((((..((((........))))...))))))))))...(((...((((....))))..))).))).))))...........(((.....)))...-.......... ( -41.10) >DroSec_CAF1 14790 119 + 1 GUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACUA-AGGCACCCAGCCACCCCCAUCAUGCC ((((.(((((((((((((..((((........))))...))))))))))...(((...((((....))))..))).))).)))).........-.(((.....))).............. ( -41.10) >DroSim_CAF1 36792 119 + 1 GUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACUA-AGGCACCCAGCCACCCCCAUCAUGCC ((((.(((((((((((((..((((........))))...))))))))))...(((...((((....))))..))).))).)))).........-.(((.....))).............. ( -41.10) >DroEre_CAF1 10647 116 + 1 GUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACCA--GCCACCCAGCCACC--CACCAUGCC ((((.(((((((((((((..((((........))))...))))))))))...(((...((((....))))..))).))).)))).........--..............--......... ( -36.60) >DroYak_CAF1 25379 116 + 1 GUGCCGCAGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACCA--CCCACUAAGCCACC--CAUUAUGCC ((((.((.((((((((((..((((........))))...))))))))))...(((...((((....))))..)))..)).)))).........--..............--......... ( -36.00) >consensus GUGCCGCUGUGCUGCAGCUUUGGCUUAGCAGCGUCACGUGCUGCAGCGCAAACUUAACGCAUUUCCGUGCCCAAGGAGCCGCACCACCCACCA_AGGCACCCAGCCACC__CAUCAUGCC ((((.(((((((((((((..((((........))))...))))))))))...(((...((((....))))..))).))).))))...........(((.....))).............. (-36.50 = -37.30 + 0.80)

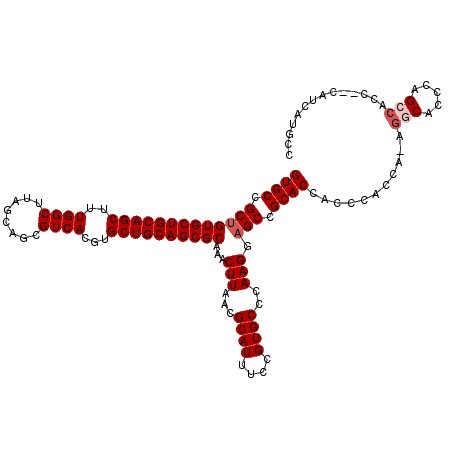
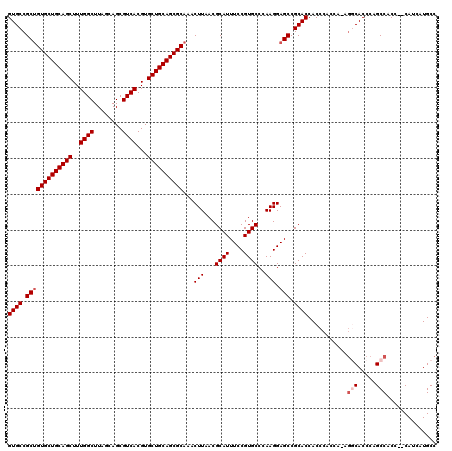
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:59 2006