
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,907,149 – 13,907,355 |
| Length | 206 |
| Max. P | 0.988265 |

| Location | 13,907,149 – 13,907,251 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.60 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13907149 102 + 22224390 CAACGCAGCAGCAGCAGCAACAUCAACUGCU---------GCAACUGCAACUGCAAUGCGGACAUCGAUGUGCGUGCUUGACCAAAAAGCAAAAAUAAAUAUAAA-AAGAAG ..(((((((((..((((((........))))---------))..))))..((((...)))).........)))))((((.......))))...............-...... ( -29.30) >DroSec_CAF1 7565 103 + 1 CAACGCAGCAGCAUCAGCAACAUCAACUGCU---------GCAACUGCAAUUCCAAUGCGGACAUCGAUGUGCGUGCUUGACCGAAAAGCAAAAAUAAAUGCAAAAAAUAAG ....((((((.((((.(((.((.....)).)---------))..(((((.......))))).....))))))).(((((.......)))))........))).......... ( -24.90) >DroSim_CAF1 29356 112 + 1 CAACGCAGCAGCAUCAGCAACAUCAACUGCUGCAACUGCUGCAACUGCAAUUGCAAUGCGGACAUCGAUGUGCGUGCUUGACCGAAAAGCAAAAAUAAAUACAAAAAAUAAG ....(((((((...(((((........)))))...)))))))...(((....)))..(((.(((....))).)))((((.......))))...................... ( -30.80) >consensus CAACGCAGCAGCAUCAGCAACAUCAACUGCU_________GCAACUGCAAUUGCAAUGCGGACAUCGAUGUGCGUGCUUGACCGAAAAGCAAAAAUAAAUACAAAAAAUAAG ..((((....((....)).(((((...(((.((....)).))).(((((.......))))).....)))))))))((((.......))))...................... (-21.93 = -22.27 + 0.33)

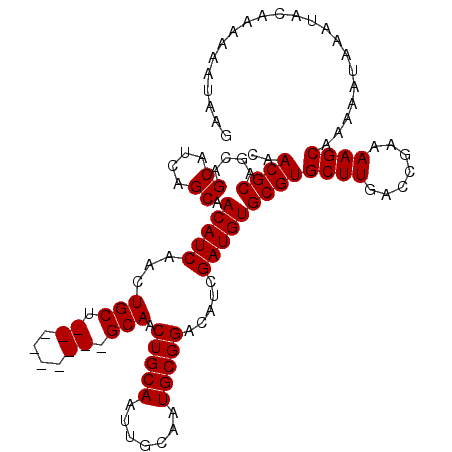

| Location | 13,907,251 – 13,907,355 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13907251 104 + 22224390 CAAUAC---AAAAAAGAAAAAAUCCCCACUCUGCUCAAUUCCGCAGCUCGCUUUAAUUGAAAGAUGUCA-AUUUGCAAACGCACAAAGUGCGUUGCGCAACAGCAACA ......---......................((((......((((((.((((((((((((......)))-)))(((....))).)))))).))))))....))))... ( -22.20) >DroSec_CAF1 7668 99 + 1 CAACAC---AAA------AAAAUCCCCACUCUGCUCAAUUCCGCAGCUCGCUUUAAUUGAAAGAUGUCAAAUUUGCAAACACACAAAGUGCGUUGCGCAACAGCAACA ......---...------.............((((......((((((.((((((..((((......))))...((......)).)))))).))))))....))))... ( -17.40) >DroSim_CAF1 29468 98 + 1 CAACAC---AAA------AAAAUGCCCACUCCGCUCAAUUCCGCAGCUCGCUUUAAUUGAAAGAUGUCA-AUUUGCAAACGCACAAAGUGCGUUGCGCAACAGCAACA ......---...------..............(((......((((((.((((((((((((......)))-)))(((....))).)))))).))))))....))).... ( -21.70) >DroEre_CAF1 3338 101 + 1 AAAUACAACAAA------AAAAUAUCCACUCUGCUCAAUUCCGCAGCGCGCUAUAAUUGAAAGAUGUCA-AUUUGCAAACGCACAAAGUGCGUUGCGCAACGGCAACA ............------.............((((......(((((((((((..((((((......)))-)))(((....)))...)))))))))))....))))... ( -26.20) >DroYak_CAF1 17716 99 + 1 AAAUACG--AAA------AAAAUGCCCACUCUGCUCAAUUCCGCAGCUCGCUUUAAUUGAAAGAUGUCA-AUUUGCAAACGCACAAAGUGCGUUGCGCAACAGCAACA .......--...------.............((((......((((((.((((((((((((......)))-)))(((....))).)))))).))))))....))))... ( -22.20) >consensus CAAUAC___AAA______AAAAUCCCCACUCUGCUCAAUUCCGCAGCUCGCUUUAAUUGAAAGAUGUCA_AUUUGCAAACGCACAAAGUGCGUUGCGCAACAGCAACA ................................((((((((..((.....))...))))))............((((.(((((((...)))))))..))))..)).... (-17.36 = -17.56 + 0.20)

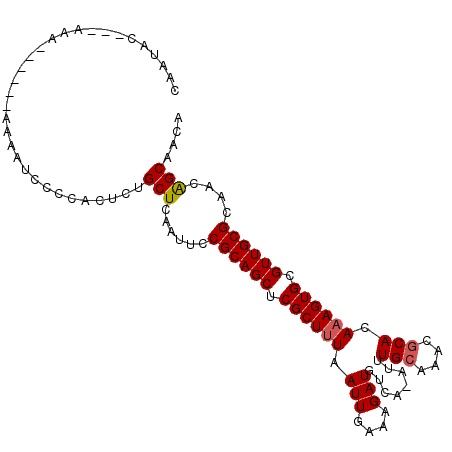

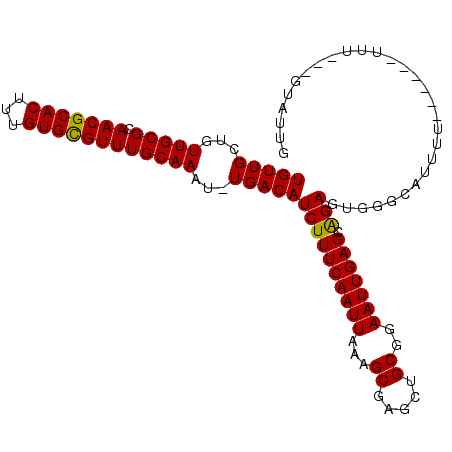
| Location | 13,907,251 – 13,907,355 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13907251 104 - 22224390 UGUUGCUGUUGCGCAACGCACUUUGUGCGUUUGCAAAU-UGACAUCUUUCAAUUAAAGCGAGCUGCGGAAUUGAGCAGAGUGGGGAUUUUUUCUUUUUU---GUAUUG .(((((......)))))((((...))))((((((.(((-(((......))))))...))))))(((((((..(((.(((((....))))))))..))))---)))... ( -27.00) >DroSec_CAF1 7668 99 - 1 UGUUGCUGUUGCGCAACGCACUUUGUGUGUUUGCAAAUUUGACAUCUUUCAAUUAAAGCGAGCUGCGGAAUUGAGCAGAGUGGGGAUUUU------UUU---GUGUUG .(((((......))))).(((((((((((((.........)))))...((((((...((.....))..))))))))))))))........------...---...... ( -23.80) >DroSim_CAF1 29468 98 - 1 UGUUGCUGUUGCGCAACGCACUUUGUGCGUUUGCAAAU-UGACAUCUUUCAAUUAAAGCGAGCUGCGGAAUUGAGCGGAGUGGGCAUUUU------UUU---GUGUUG .(((((......))))).((((((((((((((((.(((-(((......))))))...)))))).))........))))))))(((((...------...---))))). ( -27.20) >DroEre_CAF1 3338 101 - 1 UGUUGCCGUUGCGCAACGCACUUUGUGCGUUUGCAAAU-UGACAUCUUUCAAUUAUAGCGCGCUGCGGAAUUGAGCAGAGUGGAUAUUUU------UUUGUUGUAUUU .....((((.(((((((((((...))))))).((.(((-(((......))))))...)))))).)))).....(((((((.........)------))))))...... ( -29.30) >DroYak_CAF1 17716 99 - 1 UGUUGCUGUUGCGCAACGCACUUUGUGCGUUUGCAAAU-UGACAUCUUUCAAUUAAAGCGAGCUGCGGAAUUGAGCAGAGUGGGCAUUUU------UUU--CGUAUUU .(((((......)))))((((...))))((((((.(((-(((......))))))...))))))(((((((..((((.......)).))..------)))--))))... ( -27.30) >consensus UGUUGCUGUUGCGCAACGCACUUUGUGCGUUUGCAAAU_UGACAUCUUUCAAUUAAAGCGAGCUGCGGAAUUGAGCAGAGUGGGCAUUUU______UUU___GUAUUG (((((...(((((.(((((((...))))))))))))...)))))((((((((((...((.....))..))))))).)))............................. (-24.94 = -24.62 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:52 2006