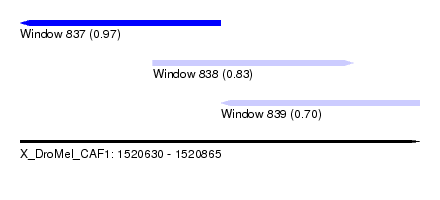
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,520,630 – 1,520,865 |
| Length | 235 |
| Max. P | 0.973351 |

| Location | 1,520,630 – 1,520,748 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.74 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1520630 118 - 22224390 CAUUAGGGUUAGUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUU--GUUAUUUGUGCCUUUUUUUUAUUUACUAACGUGUAAUUCUUUGC .((((.((((((((((((((((.((((((.(((..((((((((........)))))))).((((....))))..--.))).))))))...))))))).)))))))).).))))....... ( -26.90) >DroSec_CAF1 10050 114 - 1 CAAUAGGGUUAGUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUU--GUUAUCUGUGCCUU----UUAUUUACUAACGUGUAAUUCUUUGC .......((((((((...((((.(((((..(((..((((((((........)))))))).((((....))))..--.)))..))))).))----))..)))))))).............. ( -24.70) >DroSim_CAF1 11323 114 - 1 CAAUAGGGUUAGUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUU--GUUAUCUGUGCCUU----UUAUUUACUAACGUGUAAUUCUUUGC .......((((((((...((((.(((((..(((..((((((((........)))))))).((((....))))..--.)))..))))).))----))..)))))))).............. ( -24.70) >DroEre_CAF1 10156 114 - 1 CAAUAGGGUUAGUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUU--GUUAUUUGUGCCUU----UUAUUUACUAACGUGUAAUUCUUUGC .......((((((((...((((.((((((.(((..((((((((........)))))))).((((....))))..--.))).)))))).))----))..)))))))).............. ( -25.90) >DroYak_CAF1 11646 116 - 1 CAAUAGGGUUAAUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUUGUGUUAUUUGUGCCUU----UUAUUUACUAACGUGUAAUUCUUUGC ....(((((.(((((((.((..(((((((.....)))))))((((((((....))))))))...........)).)))))))...)))))----.....(((....)))........... ( -22.60) >consensus CAAUAGGGUUAGUAAUAGAAAAUGUACAAUUAGUUUGUAUACAUGUAUGUUUGUAUGCAUGAAAUGAGUUUCUU__GUUAUUUGUGCCUU____UUAUUUACUAACGUGUAAUUCUUUGC ....(((((.(((((((((((.(((((((.....)))))))((((((((....)))))))).......)))))...))))))...))))).........(((....)))........... (-21.86 = -22.10 + 0.24)

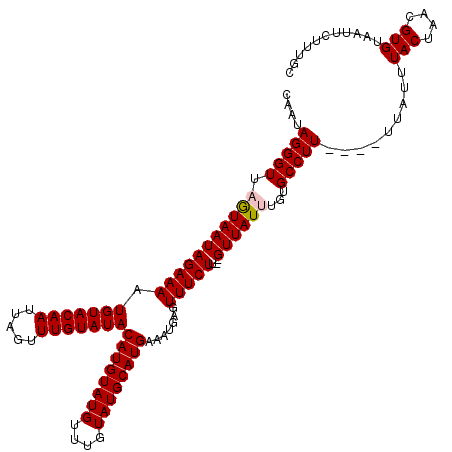
| Location | 1,520,708 – 1,520,826 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1520708 118 + 22224390 AUACAAACUAAUUGUACAUUUUCUAUUACUAACCCUAAUGAAAAUCAAACCUCAUCCCCUGUCAGCAUUAUUCACGCA-ACGCAAAACGGGGAUGCAGUGAAGAUCGAA-AACGCAUAAA .(((((.....))))).....(((.(((((........((.....)).....((((((((((..((.........)).-..)))....))))))).)))))))).....-.......... ( -17.10) >DroSec_CAF1 10124 117 + 1 AUACAAACUAAUUGUACAUUUUCUAUUACUAACCCUAUUGAAAAUCAAACCACAUCCCCUGUCAGCAUUAUUCACGCA-ACGCAAAACGGGGAUGC-GUGUAGAUCGAA-AACGCAUAAA .(((((.....)))))..(((((.(((..........(((.....)))(((.((((((((((..((.........)).-..)))....))))))).-).)).))).)))-))........ ( -18.30) >DroSim_CAF1 11397 117 + 1 AUACAAACUAAUUGUACAUUUUCUAUUACUAACCCUAUUGAAAAUCAAACCACAUCCCCUGUCAGCAUUAUUCACGCA-ACGCAAAACGGGGAUGC-GUGAAGAUCGAA-AACGCAUAAA .(((((.....))))).((((((................))))))...........((((((..((............-..))...))))))((((-((..........-.))))))... ( -17.93) >DroEre_CAF1 10230 119 + 1 AUACAAACUAAUUGUACAUUUUCUAUUACUAACCCUAUUGAAAAUCAAACAACAUCCCCUGUCAGCAUUAUUCACGCA-AGGCAAAACGGGGAUGCAGUAAAGAUCGAAAAACACAUAAA .(((((.....))))).....(((.(((((.......(((.....)))....(((((((((((.((.........)).-.))))....))))))).))))))))................ ( -20.40) >DroYak_CAF1 11722 120 + 1 AUACAAACUAAUUGUACAUUUUCUAUUAUUAACCCUAUUGAAAAUCAAACAACAUCCCCUGUCAGCAUUAUUCACGCAAAGGCAAAUCGGGGAUGCAGAGAAGAUCGAAAAACACAUAAA .(((((.....))))).(((((((.............(((.....)))....(((((((((((.((.........))...))))....)))))))...)))))))............... ( -20.10) >consensus AUACAAACUAAUUGUACAUUUUCUAUUACUAACCCUAUUGAAAAUCAAACCACAUCCCCUGUCAGCAUUAUUCACGCA_ACGCAAAACGGGGAUGCAGUGAAGAUCGAA_AACGCAUAAA .(((((.....))))).....(((.((((........(((.....)))....(((((((((....))........((....)).....)))))))..)))))))................ (-16.02 = -16.30 + 0.28)

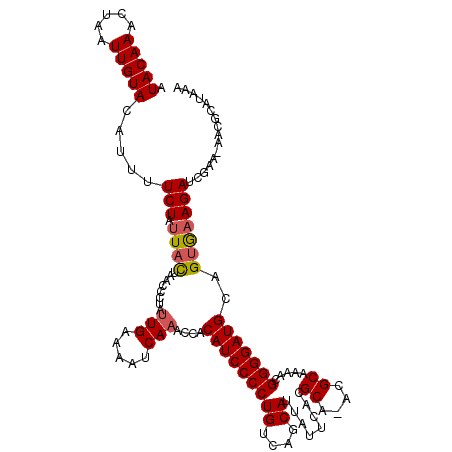
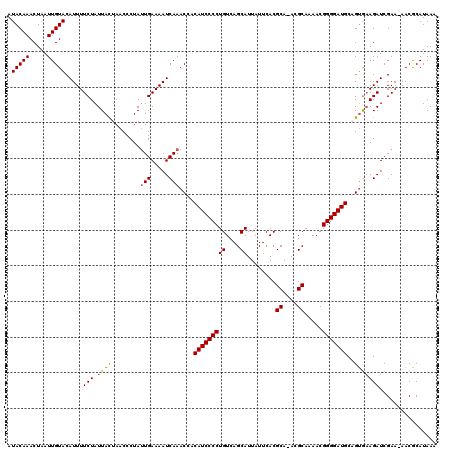
| Location | 1,520,748 – 1,520,865 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1520748 117 - 22224390 AUAAACAUCUUUUACACCU-CUCUGACUCGAUUUAUAUAUUUUAUGCGUU-UUCGAUCUUCACUGCAUCCCCGUUUUGCGU-UGCGUGAAUAAUGCUGACAGGGGAUGAGGUUUGAUUUU ...........(((.((((-(...((.((((..((((.....))))....-.))))))........(((((((((..((((-((......)))))).))).))))))))))).))).... ( -26.90) >DroSec_CAF1 10164 117 - 1 AUAAACAUCUUUUACACCUCCUCUGACUCGAUUUAUAUAUUUUAUGCGUU-UUCGAUCUACAC-GCAUCCCCGUUUUGCGU-UGCGUGAAUAAUGCUGACAGGGGAUGUGGUUUGAUUUU .(((((..................((.((((..((((.....))))....-.))))))....(-(((((((((((..((((-((......)))))).))).))))))))))))))..... ( -29.40) >DroSim_CAF1 11437 117 - 1 AUAAACAUCAUUUACACCUCCUCUGACUCGAUUUAUAUAUUUUAUGCGUU-UUCGAUCUUCAC-GCAUCCCCGUUUUGCGU-UGCGUGAAUAAUGCUGACAGGGGAUGUGGUUUGAUUUU ......((((....(((.(((((((..((((..((((.....))))....-.))))..(((((-(((....((.....)).-)))))))).........))))))).)))...))))... ( -30.90) >DroEre_CAF1 10270 119 - 1 AUAAACAUCUUUUACACUUCCUCUGACUCGAUUUAUAUAUUUUAUGUGUUUUUCGAUCUUUACUGCAUCCCCGUUUUGCCU-UGCGUGAAUAAUGCUGACAGGGGAUGUUGUUUGAUUUU .((((((.................((.((((...((((((...))))))...))))))......(((((((((((......-.((((.....)))).))).))))))))))))))..... ( -26.50) >DroYak_CAF1 11762 120 - 1 AUAAACAUCUUUUACACUUCCUCUGACUCGAUUUAUAUAUUUUAUGUGUUUUUCGAUCUUCUCUGCAUCCCCGAUUUGCCUUUGCGUGAAUAAUGCUGACAGGGGAUGUUGUUUGAUUUU .((((((.................((.((((...((((((...))))))...))))))......((((((((....((.(...((((.....)))).).))))))))))))))))..... ( -25.30) >consensus AUAAACAUCUUUUACACCUCCUCUGACUCGAUUUAUAUAUUUUAUGCGUU_UUCGAUCUUCACUGCAUCCCCGUUUUGCGU_UGCGUGAAUAAUGCUGACAGGGGAUGUGGUUUGAUUUU .(((((..................((.((((..((((.....))))......))))))......(((((((((.....)....((((.....)))).....)))))))).)))))..... (-20.74 = -20.94 + 0.20)

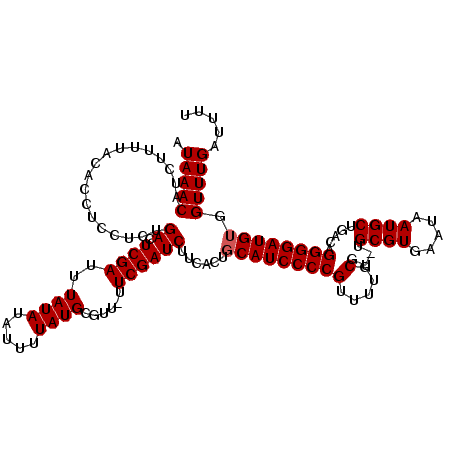
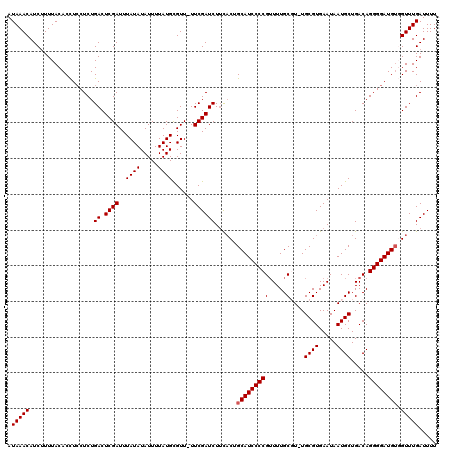
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:05 2006