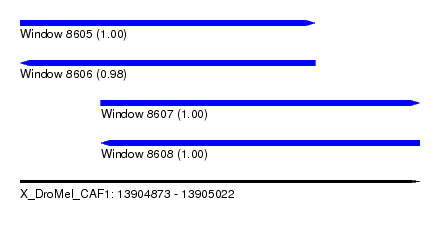
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,904,873 – 13,905,022 |
| Length | 149 |
| Max. P | 0.999863 |

| Location | 13,904,873 – 13,904,983 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13904873 110 + 22224390 ACAUUUGCUAAAGUGAUGUUCAUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCU ......(((....((((((((.((((((((((.(....).))))))..)))).))))))))(((((((....)))))))((((((.((......)).)).).))).))). ( -31.70) >DroSec_CAF1 5352 110 + 1 GCAUUUACUAAAGUGAUGUUCAUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCU ((...........((((((((.((((((((((.(....).))))))..)))).))))))))(((((((....)))))))((((((.((......)).)).).)))..)). ( -31.00) >DroSim_CAF1 27113 110 + 1 ACAUUUACUAAAGUGAUGUUCAUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCU .............((((((((.((((((((((.(....).))))))..)))).))))))))(((((((....)))))))((((((.((......)).)).).)))..... ( -29.50) >DroEre_CAF1 1168 109 + 1 ACAUUUGC-AAAGUGAUGUUCAUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUCCGAGGCAUUAAAAUGCCUCGCCCGGCCAUUUAAUUAACCAGGGGCAAGCU ...(((((-.(((((((....)))))))...((((.........(((.((.(((......((((((((....))))))))))).)).))).........))))))))).. ( -31.57) >DroYak_CAF1 15568 110 + 1 ACAUUUGCUAAAGUGAUGUUCGUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUGAGAGGCAUUAAAAUGCCUCGCCCGGCCAUUUAAUUAACCAGAGGCAAGUC .(((((....)))))((((((.((((((((((.(....).))))))..)))).))))))..(((((((....)))))))((((((............)).).)))..... ( -28.10) >consensus ACAUUUGCUAAAGUGAUGUUCAUUAUUUGUACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCU .............((((((((.((((((((((.(....).))))))..)))).))))))))(((((((....)))))))((((((.((......)).)).).)))..... (-27.10 = -27.90 + 0.80)

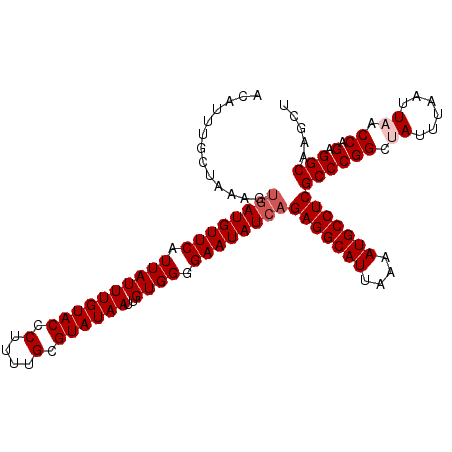

| Location | 13,904,873 – 13,904,983 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.24 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13904873 110 - 22224390 AGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAAUGAACAUCACUUUAGCAAAUGU .(((.((((..(((((......)))))))))(((((((....)))))))((((.(((..........(((.(....).)))........))).))))....)))...... ( -26.17) >DroSec_CAF1 5352 110 - 1 AGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAAUGAACAUCACUUUAGUAAAUGC .....((((..(((((......)))))))))(((((((....)))))))((((.(((..........(((.(....).)))........))).))))............. ( -23.97) >DroSim_CAF1 27113 110 - 1 AGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAAUGAACAUCACUUUAGUAAAUGU .....((((..(((((......)))))))))(((((((....)))))))((((.(((..........(((.(....).)))........))).))))............. ( -23.97) >DroEre_CAF1 1168 109 - 1 AGCUUGCCCCUGGUUAAUUAAAUGGCCGGGCGAGGCAUUUUAAUGCCUCGGAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAAUGAACAUCACUUU-GCAAAUGU .((.((((((((((((......)))))))(((((((((....)))))))((.....))............))....))))).......((.....))....-))...... ( -29.30) >DroYak_CAF1 15568 110 - 1 GACUUGCCUCUGGUUAAUUAAAUGGCCGGGCGAGGCAUUUUAAUGCCUCUCAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAACGAACAUCACUUUAGCAAAUGU .....((((..(((((......)))))))))(((((((....))))))).....................((.(((((((.............))).)))).))...... ( -24.12) >consensus AGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGUACAAAUAAUGAACAUCACUUUAGCAAAUGU .....((((..(((((......)))))))))(((((((....))))))).....................((.((((.((.((.....)).))....)))).))...... (-24.68 = -24.24 + -0.44)


| Location | 13,904,903 – 13,905,022 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -34.36 |
| Energy contribution | -35.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13904903 119 + 22224390 ACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCUGAAUGAGCCAAAUUGAACAUCCCCAAAUGGGC-AGGGGUA (((((..(((.(((.....((((((...((((((((((....)))))))((((((.((......)).)).).)))..(((.....))).....)))...)))))).))).))-)))))). ( -41.20) >DroSec_CAF1 5382 113 + 1 ACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCUGAAUGAGCCAAAUUGAACAUCCCCAAAUGGGC-------A .......(((.(((.....((((((...((((((((((....)))))))((((((.((......)).)).).)))..(((.....))).....)))...)))))).))).))-------) ( -33.60) >DroSim_CAF1 27143 119 + 1 ACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCUGAAUGAGCCAAAUUGAACAUCCCCAAAUGGGC-AGGGGCA .((((..(((.(((.....((((((...((((((((((....)))))))((((((.((......)).)).).)))..(((.....))).....)))...)))))).))).))-))))).. ( -39.70) >DroEre_CAF1 1197 119 + 1 ACCCUUUUGCGUAUAAUUGUGGGGAAUAUCCGAGGCAUUAAAAUGCCUCGCCCGGCCAUUUAAUUAACCAGGGGCAAGCUGAAUGAGCCAAAUUGAACAUCCCCAAAUGGGC-AGGGGCA .((((..(((.(((.....((((((.....((((((((....))))))))...(((((((((.((..(....)..))..)))))).)))..........)))))).))).))-))))).. ( -43.30) >DroYak_CAF1 15598 120 + 1 ACCCUUUUGCGUAUAAUUGUGGGGAAUAUGAGAGGCAUUAAAAUGCCUCGCCCGGCCAUUUAAUUAACCAGAGGCAAGUCGAAUGAGCCAAAUUGAACAUCCCCAAAUGGUUGAGGGGUA (((((((.((.(((.....((((((......(((((((....)))))))....(((((((..(((..((...))..)))..)))).)))..........)))))).))))).))))))). ( -41.80) >consensus ACCCUUUUGCGUAUAAUUGUGGGGAAUAUCAGAGGCAUUAAAAUGCCUCGCCCGGCUAUUUAAUUAACCAGAGGCAAGCUGAAUGAGCCAAAUUGAACAUCCCCAAAUGGGC_AGGGGCA .((((...((.(((.....((((((......(((((((....)))))))....(((((((((.((..((...))..)).)))))).)))..........)))))).))).)).))))... (-34.36 = -35.16 + 0.80)

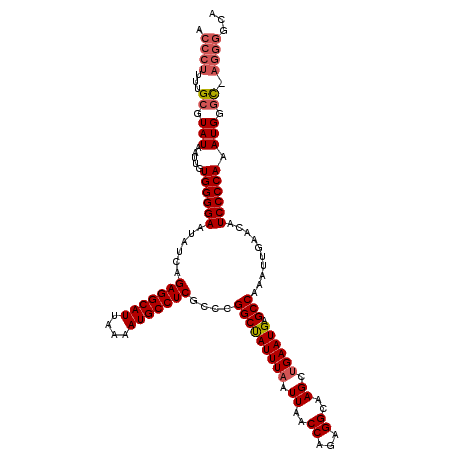
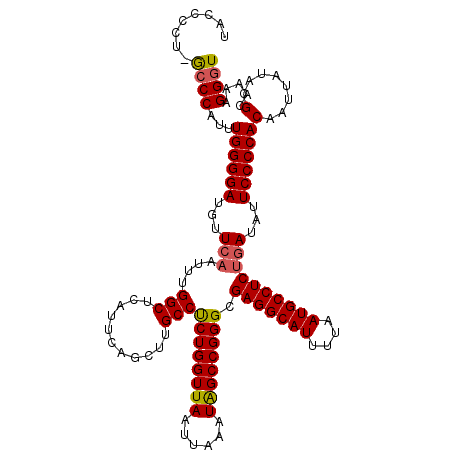
| Location | 13,904,903 – 13,905,022 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.29 |
| SVM RNA-class probability | 0.999863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13904903 119 - 22224390 UACCCCU-GCCCAUUUGGGGAUGUUCAAUUUGGCUCAUUCAGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGU .((((.(-((..(((((((((...(((....((((.....)))).((((..(((((......)))))))))(((((((....))))))))))...)))))).))).....)))...)))) ( -39.90) >DroSec_CAF1 5382 113 - 1 U-------GCCCAUUUGGGGAUGUUCAAUUUGGCUCAUUCAGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGU .-------((((...((((((...(((....((((.....)))).((((..(((((......)))))))))(((((((....))))))))))...))))))(........).....)))) ( -37.70) >DroSim_CAF1 27143 119 - 1 UGCCCCU-GCCCAUUUGGGGAUGUUCAAUUUGGCUCAUUCAGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGU .((((.(-((..(((((((((...(((....((((.....)))).((((..(((((......)))))))))(((((((....))))))))))...)))))).))).....)))...)))) ( -40.20) >DroEre_CAF1 1197 119 - 1 UGCCCCU-GCCCAUUUGGGGAUGUUCAAUUUGGCUCAUUCAGCUUGCCCCUGGUUAAUUAAAUGGCCGGGCGAGGCAUUUUAAUGCCUCGGAUAUUCCCCACAAUUAUACGCAAAAGGGU .((((.(-((..((((((((((((((.....((((.....)))).((((..(((((......)))))))))(((((((....)))))))))))).)))))).))).....)))...)))) ( -43.60) >DroYak_CAF1 15598 120 - 1 UACCCCUCAACCAUUUGGGGAUGUUCAAUUUGGCUCAUUCGACUUGCCUCUGGUUAAUUAAAUGGCCGGGCGAGGCAUUUUAAUGCCUCUCAUAUUCCCCACAAUUAUACGCAAAAGGGU ...((((........((((((.......((((((.((((..(...(((...)))...)..)))))))))).(((((((....)))))))......))))))(........)....)))). ( -33.50) >consensus UACCCCU_GCCCAUUUGGGGAUGUUCAAUUUGGCUCAUUCAGCUUGCCUCUGGUUAAUUAAAUAGCCGGGCGAGGCAUUUUAAUGCCUCUGAUAUUCCCCACAAUUAUACGCAAAAGGGU ........((((...((((((...(((....(((...........)))((((((((......)))))))).(((((((....))))))))))...))))))(........).....)))) (-34.50 = -34.54 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:46 2006