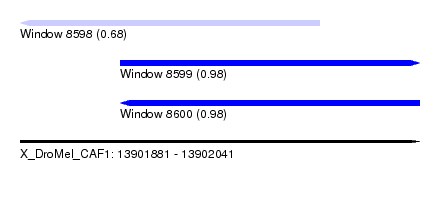
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,901,881 – 13,902,041 |
| Length | 160 |
| Max. P | 0.984725 |

| Location | 13,901,881 – 13,902,001 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -25.84 |
| Energy contribution | -28.27 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13901881 120 - 22224390 AACCCCAUUUCGAUGGGUGGAUCGGCCACUUGGCAGCCUCGAAAGGAUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGUUGCCUGGGUCUGCGUUCAAGGUACUUUUGAUUAAUUGCUC ..((((((....))))).)((((((((....))).((((.(((.(((....)))((((((....(((.(((.....))).))))))))).....))).)))).....)))))........ ( -32.00) >DroSec_CAF1 2494 120 - 1 AACCCCAUUUCGAUGGGUGGAGCGGCCACUUGGCAGCCUCGAAAGGGUUCCUCCGCCCAGAACUGUAUUUACAUUUUAGUUGCCUGGGUCUGUGUUCCACGGACUUUUGAUUAAUUGCUC .....((.(((..(((((((((.((((....)))((((.(....)))))))))))))))))).))..........(((((((...(((((((((...))))))))).)))))))...... ( -39.40) >DroSim_CAF1 24269 109 - 1 AACCCCAUUUCGAUGGGUGGAGCGG-----------CCUCGAAAGGGUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGUGGCCUGGGUCUGUGUUCCACGGACUUUUGAUUAAUUGCUC .............(((((((((..(-----------((.(....))))..))))))))).....(((.((((((....))).(..(((((((((...)))))))))..)..))).))).. ( -38.80) >DroYak_CAF1 12464 120 - 1 AACCCCAUUUCAAUGGGUGGAGUGGCCAAUUGGCAGCCACGAAAGGGUUUCUCCGCCCAGAAUUGCAUUUACGUUUUAGUUGCAUGUGUCUGCAUUCAGGGGACUUUUGAUUAAUUGCUC ..((((.......(((((((((..(((....)))((((.(....))))).)))))))))((((.(((..(((((.........)))))..))))))).)))).................. ( -44.30) >consensus AACCCCAUUUCGAUGGGUGGAGCGGCCACUUGGCAGCCUCGAAAGGGUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGUUGCCUGGGUCUGCGUUCAACGGACUUUUGAUUAAUUGCUC .....((.(((..(((((((((..(((....)))((((.(....))))).)))))))))))).))......((..(((((((...((((((((.....)))))))).))))))).))... (-25.84 = -28.27 + 2.44)



| Location | 13,901,921 – 13,902,041 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -41.19 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13901921 120 + 22224390 ACUAAAAUGUAAAUGCAGUUCUGGGCGGAGAAAUCCUUUCGAGGCUGCCAAGUGGCCGAUCCACCCAUCGAAAUGGGGUUUACUGCACGGUGGCAGCUCAACUGUAAUUACACUUUACUA ..((((.(((((.(((((((..(((.(((....))).)))(((.((((((.(((.(.......(((((....))))).......))))..)))))))))))))))).))))).))))... ( -42.64) >DroSec_CAF1 2534 120 + 1 ACUAAAAUGUAAAUACAGUUCUGGGCGGAGGAACCCUUUCGAGGCUGCCAAGUGGCCGCUCCACCCAUCGAAAUGGGGUUUCCUGCGCCGUGGCAGCUCAACUGUAAUUACACUUUACUA ..((((.(((((.((((((..((((((.(((((((((((((((((.(((....))).))).......)))))..)))).))))).)((....)).))))))))))).))))).))))... ( -47.01) >DroSim_CAF1 24309 109 + 1 ACUAAAAUGUAAAUGCAGUUCUGGGCGGAGAAACCCUUUCGAGG-----------CCGCUCCACCCAUCGAAAUGGGGUUUCCUGCGCCGUGGCAGCUCAACUGUAAUUACACUUUACUA ..((((.(((((.(((((((..(.((((.(((((((((((((((-----------.........)).)))))..))))))))))))((....))..)..))))))).))))).))))... ( -38.90) >DroYak_CAF1 12504 120 + 1 ACUAAAACGUAAAUGCAAUUCUGGGCGGAGAAACCCUUUCGUGGCUGCCAAUUGGCCACUCCACCCAUUGAAAUGGGGUUUCCUGCGCCGUGGCAGCUCAACUGUAAUUACACUUUACUA ........((((.((((.......((((((......))))))((((((((...(((((.....(((((....)))))......)).))).))))))))....)))).))))......... ( -36.20) >consensus ACUAAAAUGUAAAUGCAGUUCUGGGCGGAGAAACCCUUUCGAGGCUGCCAAGUGGCCGCUCCACCCAUCGAAAUGGGGUUUCCUGCGCCGUGGCAGCUCAACUGUAAUUACACUUUACUA ..((((.(((((.((((((..((((((((((..((.(((((((((((.....)))))..........)))))).))..)))))(((......)))))))))))))).))))).))))... (-33.94 = -34.38 + 0.44)

| Location | 13,901,921 – 13,902,041 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -45.02 |
| Consensus MFE | -36.60 |
| Energy contribution | -37.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13901921 120 - 22224390 UAGUAAAGUGUAAUUACAGUUGAGCUGCCACCGUGCAGUAAACCCCAUUUCGAUGGGUGGAUCGGCCACUUGGCAGCCUCGAAAGGAUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGU ...((((((((((...(((((..(((((......)))))..............((((((((...(((....)))....((....)).....)))))))).)))))...)))))))))).. ( -40.00) >DroSec_CAF1 2534 120 - 1 UAGUAAAGUGUAAUUACAGUUGAGCUGCCACGGCGCAGGAAACCCCAUUUCGAUGGGUGGAGCGGCCACUUGGCAGCCUCGAAAGGGUUCCUCCGCCCAGAACUGUAUUUACAUUUUAGU ...((((((((((.((((((((((((((((.(((((.(....)(((((....)))))....)).)))...)))))).)))....((((......))))..))))))).)))))))))).. ( -51.20) >DroSim_CAF1 24309 109 - 1 UAGUAAAGUGUAAUUACAGUUGAGCUGCCACGGCGCAGGAAACCCCAUUUCGAUGGGUGGAGCGG-----------CCUCGAAAGGGUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGU ...((((((((((...(((((((((.((....)))).((.....))...))..(((((((((..(-----------((.(....))))..))))))))).)))))...)))))))))).. ( -44.90) >DroYak_CAF1 12504 120 - 1 UAGUAAAGUGUAAUUACAGUUGAGCUGCCACGGCGCAGGAAACCCCAUUUCAAUGGGUGGAGUGGCCAAUUGGCAGCCACGAAAGGGUUUCUCCGCCCAGAAUUGCAUUUACGUUUUAGU ..(((((.(((((((...(((((((.((....)))).((.....))...)))))((((((((..(((....)))((((.(....))))).))))))))..))))))))))))........ ( -44.00) >consensus UAGUAAAGUGUAAUUACAGUUGAGCUGCCACGGCGCAGGAAACCCCAUUUCGAUGGGUGGAGCGGCCACUUGGCAGCCUCGAAAGGGUUUCUCCGCCCAGAACUGCAUUUACAUUUUAGU ...((((((((((...(((((...((((......))))...............(((((((((..(((....)))((((.(....))))).))))))))).)))))...)))))))))).. (-36.60 = -37.60 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:39 2006