
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,860,503 – 13,860,653 |
| Length | 150 |
| Max. P | 0.631476 |

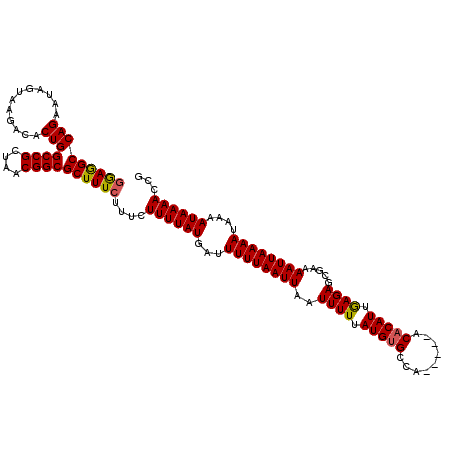
| Location | 13,860,503 – 13,860,618 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13860503 115 - 22224390 GGAAGCCAGAAUAGUAAGACACUGGCCGCUAACGGCGCUUUCUUUCUUUUAUGAUUUUUAAUUAAUUUUUAUGUGCCA-----ACACAUUGAGAGCGAAAAUUAAAAUAAAAUAAAACCG ((((((.....((((.....))))((((....))))))))))....((((((...((((((((..((((.(((((...-----.))))).)))).....))))))))....))))))... ( -24.10) >DroSec_CAF1 8137 118 - 1 GAAGGCCAGAAUAGCAAGACACUGGCCGCUAACGGCGCUUUCUUUCUUUUAUGAUUUUUAAUUAAUUUUUAUGCGCCGUGUUGGCACAU-AAGAGC-AAAAUUAAAAUAAAAUAAAACCG (((((((((............)))((((....))))))))))....((((((...((((((((..((((((((.(((.....))).)))-))))).-..))))))))....))))))... ( -30.20) >DroYak_CAF1 10736 111 - 1 GGAGGCCAGAAUAGUAAGACACUGGCCGCUAACGGCGCUUU----CUUUUAUGAUUUUUAAUUAAUUUUUAUGUGCCA-----ACACAUUGAGAGCGAAAAUUAAAAUAAAAUAAAACCA ((.((((((............))))))........((((((----(.....((((.....))))......(((((...-----.))))).)))))))....................)). ( -24.10) >consensus GGAGGCCAGAAUAGUAAGACACUGGCCGCUAACGGCGCUUUCUUUCUUUUAUGAUUUUUAAUUAAUUUUUAUGUGCCA_____ACACAUUGAGAGCGAAAAUUAAAAUAAAAUAAAACCG (((((((((............)))((((....))))))))))....((((((...((((((((..((((.(((((.........))))).)))).....))))))))....))))))... (-20.66 = -20.67 + 0.01)

| Location | 13,860,540 – 13,860,653 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13860540 113 + 22224390 --UGGCACAUAAAAAUUAAUUAAAAAUCAUAAAAGAAAGAAAGCGCCGUUAGCGGCCAGUGUCUUACUAUUCUGGCUUCCCACAGGGGGGAGGGGGUGGAGUGGGGUGGCAUUGG----- --..........................................((((....))))(((((((...((((((((.((((((......))))..)).))))))))...))))))).----- ( -32.00) >DroSec_CAF1 8175 108 + 1 CACGGCGCAUAAAAAUUAAUUAAAAAUCAUAAAAGAAAGAAAGCGCCGUUAGCGGCCAGUGUCUUGCUAUUCUGGCCUUCAACGGG-------GGGUGGAGUGUGGUGGCAUUGG----- .(((((((..................................)))))))......(((((((((..(.((((((.((((.....))-------)).)))))))..).))))))))----- ( -34.08) >DroEre_CAF1 8430 106 + 1 --UGCCACAUAAAAAUUAAUUAAAAAUCAUAAAAG----AAAGCGCCGUUAGCGGACAGUGUCUUACUAUUCUGGCCUCCCA-GGG-------GGGUGAAGUGGGGUGGCAUUGGGGGCG --.(((.............................----......(((....))).(((((((...(((((.(.((((((..-.))-------)))).))))))...)))))))..))). ( -30.50) >DroYak_CAF1 10773 106 + 1 --UGGCACAUAAAAAUUAAUUAAAAAUCAUAAAAG----AAAGCGCCGUUAGCGGCCAGUGUCUUACUAUUCUGGCCUCCCA-GGG-------GGGUGGAGUGGGGUGGCAUUGGGGGCG --.................................----...((((((....))))(((((((...((((((((.((((...-.))-------)).))))))))...)))))))...)). ( -35.30) >consensus __UGGCACAUAAAAAUUAAUUAAAAAUCAUAAAAG____AAAGCGCCGUUAGCGGCCAGUGUCUUACUAUUCUGGCCUCCCA_GGG_______GGGUGGAGUGGGGUGGCAUUGG_____ ............................................((((....))))(((((((...((((((((.((.((....)).......)).))))))))...)))))))...... (-21.80 = -22.18 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:14 2006