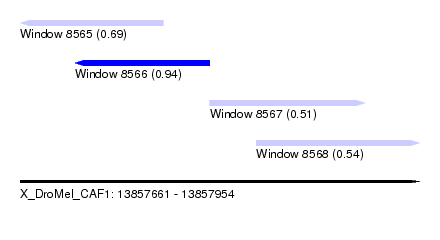
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,857,661 – 13,857,954 |
| Length | 293 |
| Max. P | 0.936026 |

| Location | 13,857,661 – 13,857,766 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13857661 105 - 22224390 CUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCGAGCAGGGAACAAACAAAAGUAAAACAAAAAAUUCGCAAACGGCCGGCGAACGGU ...(((.(((((((..((((.....))))...........(((((((.....)).(....)......................)))))...)))))))...))). ( -24.80) >DroSec_CAF1 5826 105 - 1 CUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGUCAAGCAGGGAACAAACAAAAGUAAAACAAAAAAUUCGCAAACGGCCGGCGAACGGU ...(((.(((((((..((((.....))))...........(((((((.....)).(....)......................)))))...)))))))...))). ( -24.80) >DroEre_CAF1 5613 104 - 1 CUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCAAGCAGAGAACAAGCAAAA-AAAAACUAAAAAUUCGCAAACGGCCGGCAAACGGU .......(((((((..((((.....))))...........(((((((.....))...(......)....-.............)))))...)))))))....... ( -21.50) >consensus CUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCAAGCAGGGAACAAACAAAAGUAAAACAAAAAAUUCGCAAACGGCCGGCGAACGGU ...(((.(((((((..((((.....))))...........(((((((.....)).(....)......................)))))...)))))))...))). (-23.50 = -23.50 + 0.00)


| Location | 13,857,701 – 13,857,800 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13857701 99 - 22224390 AAAAAAAA------AAACUAUAACAGAAUUUGCACAGCUGCUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCGAGCAGGGAACAAAC--------------- ........------..............(((((...(((((((((((....((...((((.....))))..))....))).).)))))))..)))))........--------------- ( -20.20) >DroSec_CAF1 5866 89 - 1 AAAAAAAA----------------CGAAUUUGCACAGCUGCUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGUCAAGCAGGGAACAAAC--------------- ........----------------....(((((..(((((((....)).)))))..((((.(...(((.(((......))).))).).)))))))))........--------------- ( -17.00) >DroEre_CAF1 5652 82 - 1 -----------------------CAGAAUUUGCACAGCUGCUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCAAGCAGAGAACAAGC--------------- -----------------------.....(((((...(((((((((((....((...((((.....))))..))....))).).)))))))..)))))........--------------- ( -19.90) >DroYak_CAF1 8121 120 - 1 AAAAAAAAAAUAUAAAACUAUAGCAGAAUUUGCACAGCUGCUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCAAGCAGAGAACAAGCUAAAUAAAACACAAA ....................((((....(((((...(((((((((((....((...((((.....))))..))....))).).)))))))..)))))......))))............. ( -23.60) >consensus AAAAAAAA_______________CAGAAUUUGCACAGCUGCUCCUGAGCCGGCUAAUUGAGGAAAUCGAAUGUAAAAUUAUGUGAGCGGCAAGCAGAGAACAAAC_______________ ............................(((((...(((((((((((....((...((((.....))))..))....))).).)))))))..)))))....................... (-18.40 = -18.40 + 0.00)



| Location | 13,857,800 – 13,857,914 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.45 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.95 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13857800 114 + 22224390 UUG------UUUUUAUAUUGCGCAUUCGGUUGAUUUCGCGCGCUGUAUCUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCAAAUUUAGGUCAAAGUCUCUA ...------.........((((...(..((((((((...(((((((.((........)))))))))...))))))))..)....))))((((((.((((.......)))))))))).... ( -34.90) >DroSec_CAF1 5955 113 + 1 GUG------UUUUUUUG-UGCGCAUUCGGUUUAUUUCGCGCGCUGUAUCUGCGCGAUGAGCAGCGCUAAAAGUCAAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUA (((------((((((((-.((((.....(((((..(((((((.......)))))))))))).))))))))))..........)))))(((((((.((((.......)))))))))))... ( -34.80) >DroEre_CAF1 5734 102 + 1 ------------------UUCGGAUUCGGUUUAUUUCGCGCGCUGUAUCUAUGCGAUGAGCAGAGAAAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUA ------------------....((((((..((.((((.(..(((.((((.....))))))).).)))).))..)))...)))...(.(((((((.((((.......)))))))))))).. ( -26.10) >DroYak_CAF1 8241 120 + 1 UUGGUUUUUUUUUUGUAUUUCGCAUUCGGUUUAUUUCGCGCGCUGUAUUUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGCCGCUA ((((((..((..((......(((.....(((((..(((((((.......)))))))))))).))).....))..))..)))))).((.((((((.((((.......)))))))))))).. ( -31.80) >consensus UUG______UUUUU_UA_UGCGCAUUCGGUUUAUUUCGCGCGCUGUAUCUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUA ..................((((...(..(((.((((...(((((((.((........)))))))))...)))).)))..)....))))((((((.((((.......)))))))))).... (-24.51 = -24.95 + 0.44)

| Location | 13,857,834 – 13,857,954 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -33.76 |
| Energy contribution | -34.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13857834 120 + 22224390 CGCUGUAUCUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCAAAUUUAGGUCAAAGUCUCUAGUUUGUGCGGUCGUUUAUCUCGCUCGGAAUGGCGAAAAAC ((((((.((((.((((((((((.(((.....((.((....)).))((((((((((((((.......)))))))......)))))))))).).)))))..)))).))))))))))...... ( -37.20) >DroSec_CAF1 5988 120 + 1 CGCUGUAUCUGCGCGAUGAGCAGCGCUAAAAGUCAAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUAGUCUGUGCGGUCGCUUAUCUCGCUCGGAAUGGCGAAAAAC ((((((.((((((.((((((((.(((.....((..........))((((((((((((((.......)))))))......)))))))))).).))))))).)))..)))))))))...... ( -40.40) >DroEre_CAF1 5756 120 + 1 CGCUGUAUCUAUGCGAUGAGCAGAGAAAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUAGUUUGUGCGGUCGUUUAUCUCGCUCGGAAUGGCGAAAAGC (((((((((.....)))((((.((((.(((...(((((.((.(((..(((((((.((((.......)))))))))))...))).))..)))))))).))))))))...))))))...... ( -34.60) >DroYak_CAF1 8281 120 + 1 CGCUGUAUUUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGCCGCUAGUUUGUGCGGUCGUUUAUCUCGCUCGGAAUGGCGAAAAAC ((((((.((((.((((((((((.(((((((.((.((....)).))((.((((((.((((.......))))))))))))...)))).))).).)))))..)))).))))))))))...... ( -38.70) >consensus CGCUGUAUCUGUGCGAUGAGCAGCGCUAAAAGUCGAUUAAUCAACGCAGGCUUUUGGCCGAAUUUAGGUCAAAGUCUCUAGUUUGUGCGGUCGUUUAUCUCGCUCGGAAUGGCGAAAAAC ((((((.((((.((((((((((.(((.....((.((....)).))((((((((((((((.......)))))))......)))))))))).).)))))..)))).))))))))))...... (-33.76 = -34.20 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:10 2006