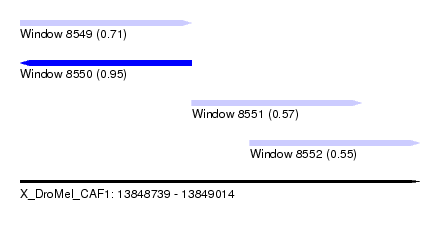
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,848,739 – 13,849,014 |
| Length | 275 |
| Max. P | 0.952163 |

| Location | 13,848,739 – 13,848,857 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -31.10 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13848739 118 + 22224390 GUAAACCAACUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGACCGUUCUAUGCUUUUUUU--UAAUAUUCAUAUUUU ..........((((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))))...................--............... ( -31.50) >DroSim_CAF1 2641 118 + 1 UUUAAACAACUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAUCCUUCUAUACUUUUUUU--CAAUAUUAAUAUUUU ..........((((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))))...................--............... ( -31.50) >DroYak_CAF1 75 119 + 1 GUAAAUCAACUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAAGUUUAACACUUUUUUUUUAAGUGUUUAUAUUU- ..........((((((((..((((........)))).((((((....).))))).....(((((.......))))))))))))).((((..(((((((.......)))))))...))))- ( -37.20) >consensus GUAAAACAACUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAACGUUCUAUACUUUUUUU__AAAUAUUAAUAUUUU ..........((((((((..((((........))))...(((((.((((((.(.......).)))))))))))...)))))))).................................... (-31.10 = -31.10 + -0.00)



| Location | 13,848,739 – 13,848,857 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13848739 118 - 22224390 AAAAUAUGAAUAUUA--AAAAAAAGCAUAGAACGGUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAGUUGGUUUAC ...............--..........((((.(((..(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))..))).)))). ( -30.90) >DroSim_CAF1 2641 118 - 1 AAAAUAUUAAUAUUG--AAAAAAAGUAUAGAAGGAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAGUUGUUUAAA .........(((((.--......))))).(((.(((.(((((((.((((.......))))......((((((.....)))))).((((........))))..))))))).))).)))... ( -31.70) >DroYak_CAF1 75 119 - 1 -AAAUAUAAACACUUAAAAAAAAAGUGUUAAACUUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAGUUGAUUUAC -.......(((((((.......)))))))........(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))........... ( -34.10) >consensus AAAAUAUAAAUAUUA__AAAAAAAGUAUAGAACGAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAGUUGAUUUAC .....................................(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))........... (-28.60 = -28.60 + -0.00)



| Location | 13,848,857 – 13,848,974 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.31 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -15.72 |
| Energy contribution | -17.95 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13848857 117 + 22224390 UCCUUGAGCUAUGAAUAUUACAGCUUUCAUUAAUUGGCCAAGUCAAUUGCUGAAAA---AAAUAUUUAUUAGUUCUUUAAGGAACUAGAAGCUUAAAUAGUGUAUUGGGCUUGCGUAAGG .(((((.((..((((((((.....(((((.((((((((...)))))))).))))).---.))))))))((((((((....))))))))((((((((........)))))))))).))))) ( -31.10) >DroSim_CAF1 2759 117 + 1 UCCUUGAGCUAUGAAUAUUACAGCUUUUUUUAAUUGGCCAAGUCAAUUGUUGCCAC---AAAUAUUUAUAAGUUCUUUAAGGAACUAGAAGCCUAAAUAGUGUAUUGGGCUUGCCAAAUA (((((((((((((((((((...((......((((((((...))))))))..))...---.))))))))).)))))...))))).....((((((((........))))))))........ ( -28.80) >DroYak_CAF1 194 118 + 1 UGCUUGAGCUCAGAAUAUUACAGAUUUUAUUAAUUGGUCAAGCCUAUUGCUGCUAAAAAAGAGAGUUAU--GUUCGCUGAGAAGCAAGAAUCUUAACUAGCGUAUUGGGCGCCUAUAAUA .((((((.(....((((..........))))....).))))(((((.(((.((((...(((((((....--.)))(((....))).....))))...))))))).)))))))........ ( -25.00) >consensus UCCUUGAGCUAUGAAUAUUACAGCUUUUAUUAAUUGGCCAAGUCAAUUGCUGCAAA___AAAUAUUUAU_AGUUCUUUAAGGAACUAGAAGCUUAAAUAGUGUAUUGGGCUUGCAUAAUA (((((((((((((((((((...........((((((((...))))))))...........))))))))).)))))...))))).....((((((((........))))))))........ (-15.72 = -17.95 + 2.23)


| Location | 13,848,897 – 13,849,014 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -21.31 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13848897 117 + 22224390 AGUCAAUUGCUGAAAA---AAAUAUUUAUUAGUUCUUUAAGGAACUAGAAGCUUAAAUAGUGUAUUGGGCUUGCGUAAGGACAAGUAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACU (((((..(((((....---.........((((((((....))))))))((((((((........)))))))).............)))))(((.(((((....))))).)))...))))) ( -30.10) >DroSim_CAF1 2799 117 + 1 AGUCAAUUGUUGCCAC---AAAUAUUUAUAAGUUCUUUAAGGAACUAGAAGCCUAAAUAGUGUAUUGGGCUUGCCAAAUAACAAGUAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACU (((((..(((((((((---...........((((((....))))))..((((((((........))))))))((((....((........))..))))...)))))...))))..))))) ( -32.60) >DroYak_CAF1 234 118 + 1 AGCCUAUUGCUGCUAAAAAAGAGAGUUAU--GUUCGCUGAGAAGCAAGAAUCUUAACUAGCGUAUUGGGCGCCUAUAAUAAUAUGCAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACU .((((((((((((..........(((((.--(((((((....)))..))))..))))).((((.....))))............))))))))..(((((....)))))))))........ ( -33.20) >consensus AGUCAAUUGCUGCAAA___AAAUAUUUAU_AGUUCUUUAAGGAACUAGAAGCUUAAAUAGUGUAUUGGGCUUGCAUAAUAACAAGUAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACU ((((...((((((..................(((((....)))))...((((((((........))))))))............))))))(((.(((((....))))).)))....)))) (-21.31 = -22.77 + 1.45)

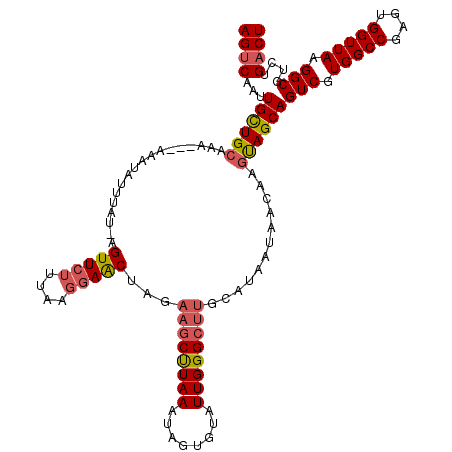
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:56 2006