| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,814,548 – 13,814,653 |
| Length | 105 |
| Max. P | 0.787782 |

| Location | 13,814,548 – 13,814,653 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13814548 105 - 22224390 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCG--CCAGCAUCAUAUUCCAUCUUCU-GGCUCUGCCGAAGACCCCUCUUCA------------ ..........(((((.(((((........((((......)))).(((....))))))))--.)))))..........(((((.-(((...))))))))..........------------ ( -24.60) >DroPse_CAF1 142259 110 - 1 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCGCGCCUGC---AUCUUCCAUCUUCC-UUC--C----UUGACCCCUCUGUGCGCCUGCUCCCC ...........((.((.(((((.......((((......))))((((...(((.....)))...))---))............-...--.----..........)))))))..))..... ( -17.90) >DroEre_CAF1 123644 103 - 1 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCG--CCAGCAUCAUCUUCCAUCUUCU-UGC--AGCAGAAGAUCCCUCUUCA------------ ........(((.((((((((.......(((((((.......((.(((....))).))..--....)))))))...........-)))--))))))))...........------------ ( -26.79) >DroYak_CAF1 133775 104 - 1 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCG--CCAGCAUCAUCUUCCAGCUUCUUUGC--AGCAGAAGACCCCUCUUCG------------ ........(((.((((((((.......(((((((.......((.(((....))).))..--....)))))))............)))--))))))))...........------------ ( -26.23) >DroAna_CAF1 132079 95 - 1 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCG--CCAGCAU---CUUCCAACUUCC-UGU--GGC---AGCCCCCCCCU-------------- ...........(((((..((.....(((.((((......))))(((((..(((...)))--..)))))---....))).....-)).--.))---)))........-------------- ( -23.20) >DroPer_CAF1 143270 106 - 1 UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCGCGCCUGC---AUCUUCCAUCUU-----C--C----UUGACCCCUCUGUGCCCCUGCUCCCC ...........((.(..(((((.......((((......))))((((...(((.....)))...))---))..........-----.--.----..........)))))..).))..... ( -15.60) >consensus UAUAUUAACUUGCUGCUGCAUAUAAUUGAUGAUGAUUAAAUCAAUGCUACCGCAAUGCG__CCAGCAU_AUCUUCCAUCUUCU_UGC__AGC__AAGACCCCUCUUCG____________ ...........((((.(((((........((((......)))).(((....))))))))...))))...................................................... (-12.07 = -12.40 + 0.33)

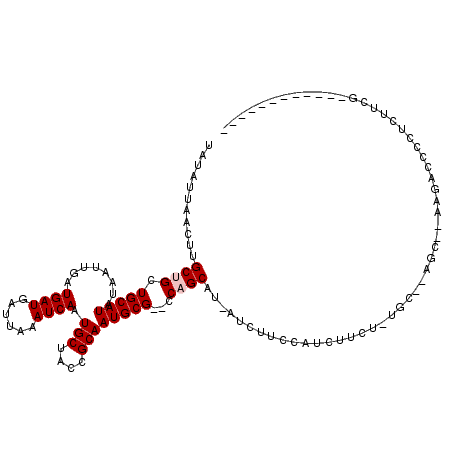
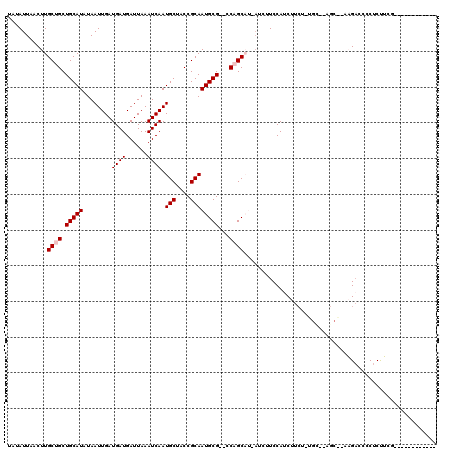
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:49 2006