| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,794,845 – 13,794,944 |
| Length | 99 |
| Max. P | 0.776119 |

| Location | 13,794,845 – 13,794,944 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -16.44 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
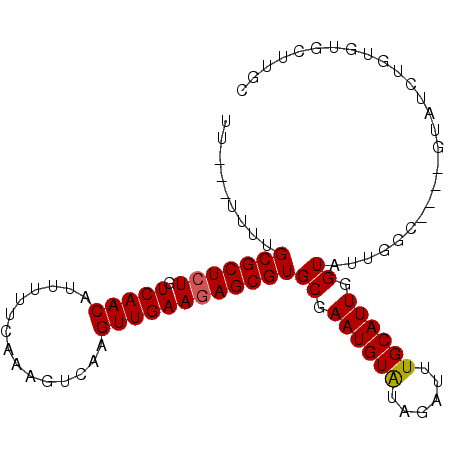
>X_DroMel_CAF1 13794845 99 + 22224390 GCAAGCACACGGAUACAGAUACCAAUACCAAUGCAAAUCUACACAUUCGCACGCUCUUCAACUUGACUUUGAAAAAUGUUGACAGAGCGCAAAAAAAAA ....((...(((((..((((..((.......))...))))....)))))...((((((((((...............))))).)))))))......... ( -16.36) >DroSec_CAF1 101589 92 + 1 GCAAACACACAGAUAC----GCCAAUACCAAUGCAAAUCUACACAUUCGCACGCUAUUCAACUUGACUUUGAAAAAUGUUGACAGAGCGCAAAU---AA ((........((((..----((..........))..))))............(((..(((((...............)))))...)))))....---.. ( -11.46) >DroEre_CAF1 103850 91 + 1 GCAAGCACACAGAUAC----GCCAAUACCAAUGCAUAUCUAUACAUUCGCACGCUCUUCAACUUGACUUUGAAAAAUGUUGACAGAGCGCGAA----AA ..........(((((.----((..........)).))))).....(((((..((((((((((...............))))).))))))))))----.. ( -20.66) >DroYak_CAF1 113142 92 + 1 GCUAGCACACAGAUAG----GCCAAUACCAAUGCAAAUCUAUACAUUCGCACGCUCUUCAACUUGACUUUGAAAAAUGUUGACAGAGCGCAAAA---AA ....((....((((..----((..........))..))))............((((((((((...............))))).)))))))....---.. ( -17.26) >consensus GCAAGCACACAGAUAC____GCCAAUACCAAUGCAAAUCUACACAUUCGCACGCUCUUCAACUUGACUUUGAAAAAUGUUGACAGAGCGCAAAA___AA ((.............................(((.(((......))).))).((((((((((...............))))).)))))))......... (-13.69 = -14.18 + 0.50)


| Location | 13,794,845 – 13,794,944 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.81 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13794845 99 - 22224390 UUUUUUUUUGCGCUCUGUCAACAUUUUUCAAAGUCAAGUUGAAGAGCGUGCGAAUGUGUAGAUUUGCAUUGGUAUUGGUAUCUGUAUCCGUGUGCUUGC .........(((((((.(((((...............))))))))))))((((((......))))))...((((((((.........))).)))))... ( -22.96) >DroSec_CAF1 101589 92 - 1 UU---AUUUGCGCUCUGUCAACAUUUUUCAAAGUCAAGUUGAAUAGCGUGCGAAUGUGUAGAUUUGCAUUGGUAUUGGC----GUAUCUGUGUGUUUGC ..---....(((((...(((((...............)))))..)))))(((((((..(((((.(((..........))----).)))))..))))))) ( -25.16) >DroEre_CAF1 103850 91 - 1 UU----UUCGCGCUCUGUCAACAUUUUUCAAAGUCAAGUUGAAGAGCGUGCGAAUGUAUAGAUAUGCAUUGGUAUUGGC----GUAUCUGUGUGCUUGC ..----...(((((((.(((((...............))))))))))))((((.(..((((((((((..........))----))))))))..).)))) ( -29.76) >DroYak_CAF1 113142 92 - 1 UU---UUUUGCGCUCUGUCAACAUUUUUCAAAGUCAAGUUGAAGAGCGUGCGAAUGUAUAGAUUUGCAUUGGUAUUGGC----CUAUCUGUGUGCUAGC ..---....(((((((.(((((...............))))))))))))((.((((((......)))))).)).(((((----(.......).))))). ( -24.76) >consensus UU___UUUUGCGCUCUGUCAACAUUUUUCAAAGUCAAGUUGAAGAGCGUGCGAAUGUAUAGAUUUGCAUUGGUAUUGGC____GUAUCUGUGUGCUUGC .........(((((((.(((((...............))))))))))))((.((((((......)))))).)).......................... (-19.81 = -19.81 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:44 2006