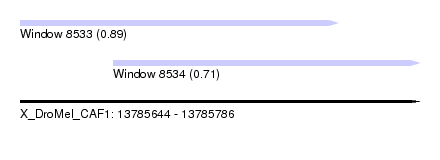
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,785,644 – 13,785,786 |
| Length | 142 |
| Max. P | 0.893018 |

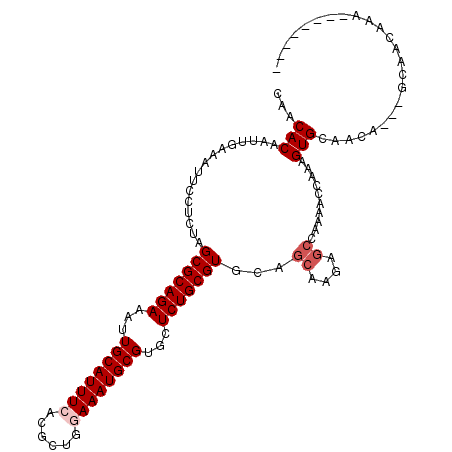
| Location | 13,785,644 – 13,785,757 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -17.28 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13785644 113 + 22224390 CU---CCCGCACCACUCAUCCCCCUUC----GGGCCACACCAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAAAGCCAAAA ..---...((((..........((...----.))..............................(((((...((((((((......))))))))...))))))))).((....))..... ( -27.20) >DroPse_CAF1 107026 104 + 1 CUUGCCCCGCG---C------CGCUCCGCGGUGGCCACACCAACACAAUUGAAAUUCCUCUGGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCUGCGUGCAGCA-------AAA ...((((((((---.------.....))))).)))........................((((((((((...((((((((.....).)))))))...))))))).)))..-------... ( -33.00) >DroEre_CAF1 94561 110 + 1 CU---CCCGCA---CUCAUCCCCCUUC----GGGCCACACCAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAAAGCCAAAA ..---...(((---(.......((...----.))..............................(((((...((((((((......))))))))...))))))))).((....))..... ( -27.60) >DroWil_CAF1 103968 85 + 1 -------------------------UC----GUCUCACACCAACACAAUUGAAAUUCCUUUCGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCCGCGUGCAGG------CAACA -------------------------..----(((((((............((((....))))(((.((....((((((((.....).)))))))..)).)))))).)))------).... ( -20.50) >DroYak_CAF1 103230 110 + 1 CU---CCCGCA---CUCAUCCCCCUGC----GGGCCACACCAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAGAGCCAAAA ..---((((((---..........)))----))).......................((((.(((((((...((((((((......))))))))...)))))))......))))...... ( -31.40) >DroMoj_CAF1 123664 90 + 1 -------GGCA-----------CUCUU----GGCACACACCAACACAAUUGAAAUUCCUUUGGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCUGCGUGCAGCA---GC----- -------.(((-----------(.(..----(((((.((.(((.....))).......((..(((..(((((...))))).)))..))..)).)))))..).))))....---..----- ( -23.00) >consensus CU___CCCGCA___C______CCCUUC____GGGCCACACCAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCUGCGUGCAGCA___GCCAAAA ........((.....................((......)).....................(((((((...(((((((........)))))))...)))))))...))........... (-17.28 = -18.12 + 0.83)

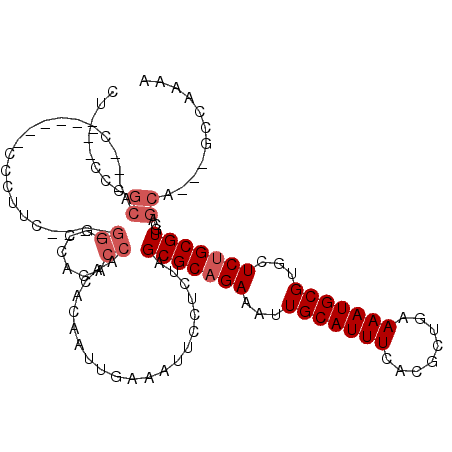
| Location | 13,785,677 – 13,785,786 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13785677 109 + 22224390 CAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAAAGCCAAAACCAAAGUGCAAUA---GCAACAAAAAAAAAAC ...(((................(((((((...((((((((......))))))))...)))))))...((....))..........))).....---................ ( -23.70) >DroPse_CAF1 107057 95 + 1 CAACACAAUUGAAAUUCCUCUGGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCUGCGUGCAGCA-------AAACCAAAGUGCAACG---ACGAUAGG-------C .....(.((((...........(((((((...((((((((.....).)))))))...)))))))...(((-------.........)))....---.)))).).-------. ( -20.20) >DroSec_CAF1 93011 101 + 1 CAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAGAGCCAAAACCAAAGUGCAACA---GCAACAAA-------- ........(((......((((.(((((((...((((((((......))))))))...)))))))......))))............(((....---))).))).-------- ( -25.20) >DroSim_CAF1 99372 101 + 1 CAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAGAGCCAAAACCAAAGUGCAACA---GCAACAAA-------- ........(((......((((.(((((((...((((((((......))))))))...)))))))......))))............(((....---))).))).-------- ( -25.20) >DroYak_CAF1 103260 101 + 1 CAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAGAGCCAAAACCAAAGUGCAACA---GCAACAAA-------- ........(((......((((.(((((((...((((((((......))))))))...)))))))......))))............(((....---))).))).-------- ( -25.20) >DroMoj_CAF1 123682 97 + 1 CAACACAAUUGAAAUUCCUUUGGCGCAGAAAUUGCAUUUCACGCUGAAAAUGCGUGCUCUGCGUGCAGCA---GC-----CUAAAGUGCAACGAUAGCAGCAGCA------- .....(((..(.....)..)))((((.....((((((((...((((....(((((((...))))))).))---))-----...)))))))).....)).))....------- ( -23.40) >consensus CAACACAAUUGAAAUUCCUCUAGCGCAGAAAUUGCAUUUCACGCUGGAAAUGCGUGCUCUGCGUGCAGCAAGAGCCAAAACCAAAGUGCAACA___GCAACAAA________ ...(((................(((((((...((((((((......))))))))...)))))))...((....))..........)))........................ (-20.50 = -21.17 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:40 2006