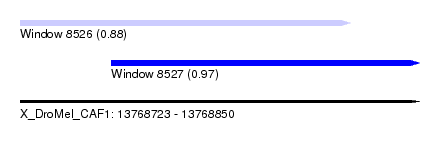
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,768,723 – 13,768,850 |
| Length | 127 |
| Max. P | 0.971497 |

| Location | 13,768,723 – 13,768,828 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -16.09 |
| Energy contribution | -15.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13768723 105 + 22224390 ACCCACACAUUGAUAAGCCCCACCAACGCCCCAC----CCCCUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGGAAGUGCACAAAUUGUUGAAAUUUUC .........(..((((..........(((.....----.........))).((((((((....((((((....))))))...)))))))).....))))..)....... ( -21.24) >DroSec_CAF1 79257 109 + 1 ACCCGCACAUUGAUAAGCCCCCUCAACGCCCCUCUCCACCCAUUUCCGCGGGCACUUUUAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC ......................(((((............((........))((((((((....((((((....))))))...)))))))).......)))))....... ( -21.40) >DroSim_CAF1 85469 109 + 1 ACCCGCACAUUGAUAAGCCCCCUCACCGCCCCCCUCCGCCCAUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC .........(..((((...................((((........))))((((((((....((((((....))))))...)))))))).....))))..)....... ( -25.90) >DroEre_CAF1 80318 99 + 1 CCCC-CCCUUAGAUGAGCUCCUUCAC-------CGC--CCCCUUCCCCGCGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC ....-................((((.-------(((--..........)))((((((((....((((((....))))))...)))))))).........))))...... ( -20.50) >DroYak_CAF1 88467 106 + 1 ACCC-CCCUUUGAUAAACCCCUUUACGCCCACCCCC--CACCUUCCCGAUGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC ....-...((..((((....................--((.(.....).))((((((((....((((((....))))))...)))))))).....))))..))...... ( -19.20) >DroAna_CAF1 86394 100 + 1 UCCCCUAGAUUAGCCAUCCCCCUCAUCCCUCCCC---------ACCCCCCGCCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC ..................................---------.......((.((((((....((((((....))))))...))))))))................... ( -13.80) >consensus ACCC_CACAUUGAUAAGCCCCCUCACCGCCCCCC_C__CCCCUUCCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUC ...................................................((((((((....((((((....))))))...))))))))................... (-16.09 = -15.98 + -0.11)

| Location | 13,768,752 – 13,768,850 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13768752 98 + 22224390 CCCAC----CCCCUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGGAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAA---CG .....----..((.......))((((((((....((((((....))))))...)))))))).....((((((....((((.....))))....)))))).---.. ( -24.80) >DroSim_CAF1 85498 102 + 1 CCCCCUCCGCCCAUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAA---CG ......((((........))))((((((((....((((((....))))))...)))))))).....((((((....((((.....))))....)))))).---.. ( -30.70) >DroEre_CAF1 80343 96 + 1 ----CGC--CCCCUUCCCCGCGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAA---CG ----(((--..........)))((((((((....((((((....))))))...)))))))).....((((((....((((.....))))....)))))).---.. ( -26.40) >DroYak_CAF1 88495 100 + 1 CACCCCC--CACCUUCCCGAUGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUGGCGAAAUCAGCAACAAA---CG .......--...........((((((((((....((((((....))))))...)))))))).))..((((((....((((.....))))....)))))).---.. ( -25.40) >DroAna_CAF1 86423 93 + 1 UCCCC---------ACCCCCCGCCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAA---CG .....---------.......((.((((((....((((((....))))))...)))))))).....((((((....((((.....))))....)))))).---.. ( -21.40) >DroPer_CAF1 92154 93 + 1 CAAAC----CCCCUC--------CCUCCUCAAACAGUUUGCUCGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAAAACCG .....----......--------............(((((....))))).................((((((....((((.....))))....))))))...... ( -12.40) >consensus CCCCC____CCCCUUCCCGCCGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUUUUCUUAGCGAAAUCAGCAACAAA___CG ......................((((((((....((((((....))))))...)))))))).....((((((....((((.....))))....))))))...... (-20.48 = -21.20 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:34 2006