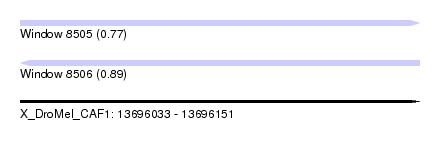
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,696,033 – 13,696,151 |
| Length | 118 |
| Max. P | 0.885415 |

| Location | 13,696,033 – 13,696,151 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
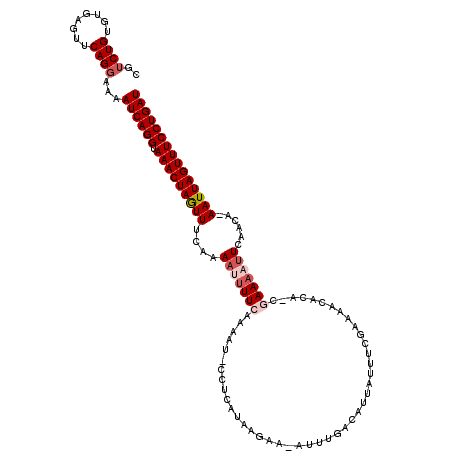
>X_DroMel_CAF1 13696033 118 + 22224390 AUCAGGAAACUAAUU-UGUUGAAUUUUCGAAAUGUUUUCGAUAUAAUGUCAAAU-UUCUUAUGAAGGCUUUGGAAAAUUUGGAAAUUAGUUUACCUGAUUUUCCUGAACCCACACAGACG (((((((((((((((-(.(..(((((((.(((.((((((.(((.(((.....))-)...)))))))))))).)))))))..))))))))))).))))))....(((........)))... ( -32.80) >DroSec_CAF1 7963 118 + 1 AUCAGGAAACUAAUU-UGUUGAAUUUUCGAAAUGUUUUCGAAAUAAUGUCAAAU-UUCUUAUGAGGAAUUUUGAAAAUUUUGAAACUAGUUUGCCUGAUUUUCCUGAACUCACACAGACG ((((((((((((.((-(...(((((((((((((.((((((((((........))-)))....))))))))))))))))))..))).)))))).))))))....(((........)))... ( -27.30) >DroSim_CAF1 15644 107 + 1 AUCAGGAAACUAAUU-UGUUGAAUUUUC-----------GAAAUAAUGUCAAAU-UUCUUAUGAGGAAUUUUGAAAAUUUUGAAACUAGUUUACCUGAUUUUCCUGAACUCACACAGACG ((((((((((((.((-(...((((((((-----------(((((..(.(((...-......))).).)))))))))))))..))).)))))).))))))....(((........)))... ( -23.70) >DroEre_CAF1 8279 101 + 1 AUCAGGAAACUAAUU-UGUGGAAUUUUUG-UGUGUUCUCGAAAUGGUGUCAGAU-----------------UUAAAGUUUUGAAACUAGUUUACCUGAUUUUGCUGAACUCACACAGACG ((((((((((...((-((.(((((.....-...))))))))).((((.(((((.-----------------.......))))).)))))))).))))))....(((........)))... ( -21.00) >DroYak_CAF1 13617 118 + 1 AUCAGGAAACUAAUUUUGUGAAUAAUUUU-UGUAUUCUCGAAAUAGUGUCAAAUGUUGUUUUGAAG-AUUUUGAAAGUUUUGAAACUAGUUUACCUGAUUUUCCUGAACUCACACAGACG ((((((((((((.(((((.....((((((-((...(((((((((((..(...)..)))))))).))-)...)))))))).))))).)))))).))))))....(((........)))... ( -25.00) >consensus AUCAGGAAACUAAUU_UGUUGAAUUUUCG_AAUGUUCUCGAAAUAAUGUCAAAU_UUCUUAUGAAG_AUUUUGAAAAUUUUGAAACUAGUUUACCUGAUUUUCCUGAACUCACACAGACG ((((((((((((.((...((((((((((............................................)))))))))).)).)))))).))))))....(((........)))... (-13.54 = -14.18 + 0.64)



| Location | 13,696,033 – 13,696,151 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13696033 118 - 22224390 CGUCUGUGUGGGUUCAGGAAAAUCAGGUAAACUAAUUUCCAAAUUUUCCAAAGCCUUCAUAAGAA-AUUUGACAUUAUAUCGAAAACAUUUCGAAAAUUCAACA-AAUUAGUUUCCUGAU ..((((........))))...((((((.((((((((((.((((((((...............)))-)))))........(((((.....))))).........)-))))))))))))))) ( -24.16) >DroSec_CAF1 7963 118 - 1 CGUCUGUGUGAGUUCAGGAAAAUCAGGCAAACUAGUUUCAAAAUUUUCAAAAUUCCUCAUAAGAA-AUUUGACAUUAUUUCGAAAACAUUUCGAAAAUUCAACA-AAUUAGUUUCCUGAU ..((((........))))...((((((.((((((((((........(((((.(((.......)))-.))))).....(((((((.....))))))).......)-))))))))))))))) ( -26.40) >DroSim_CAF1 15644 107 - 1 CGUCUGUGUGAGUUCAGGAAAAUCAGGUAAACUAGUUUCAAAAUUUUCAAAAUUCCUCAUAAGAA-AUUUGACAUUAUUUC-----------GAAAAUUCAACA-AAUUAGUUUCCUGAU ..((((........))))...((((((.((((((((((...(((((((.((((...(((.((...-.)))))....)))).-----------)))))))....)-))))))))))))))) ( -24.10) >DroEre_CAF1 8279 101 - 1 CGUCUGUGUGAGUUCAGCAAAAUCAGGUAAACUAGUUUCAAAACUUUAA-----------------AUCUGACACCAUUUCGAGAACACA-CAAAAAUUCCACA-AAUUAGUUUCCUGAU ....((((((..(((.......((((((.....((((....))))....-----------------)))))).........)))..))))-))...........-.(((((....))))) ( -14.99) >DroYak_CAF1 13617 118 - 1 CGUCUGUGUGAGUUCAGGAAAAUCAGGUAAACUAGUUUCAAAACUUUCAAAAU-CUUCAAAACAACAUUUGACACUAUUUCGAGAAUACA-AAAAUUAUUCACAAAAUUAGUUUCCUGAU ..((((........))))...((((((.((((((((((........((.((((-..(((((......)))))....)))).))(((((..-.....)))))...)))))))))))))))) ( -21.40) >consensus CGUCUGUGUGAGUUCAGGAAAAUCAGGUAAACUAGUUUCAAAAUUUUCAAAAU_CCUCAUAAGAA_AUUUGACAUUAUUUCGAAAACACA_CGAAAAUUCAACA_AAUUAGUUUCCUGAU ..((((........))))...((((((.(((((((((....(((((((............................................)))))))......))))))))))))))) (-13.26 = -14.50 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:16 2006