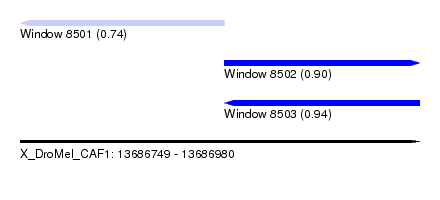
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,686,749 – 13,686,980 |
| Length | 231 |
| Max. P | 0.943453 |

| Location | 13,686,749 – 13,686,867 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13686749 118 - 22224390 CCUACAAACA--AGCUAAACAGGCAAGAAUAGUUUAUAGCGAUGGCAACCAUGGCAAGCAAUAGAGAAAUUGGACGGUGCUGGCAAUCCUGCAGCUGCUGCAUCGUCCAUCCGCUGGAAA ((........--.(((....((((.......))))...((.((((...)))).)).))).....((....((((((((((.((((..........))))))))))))))....))))... ( -32.60) >DroSim_CAF1 3945 118 - 1 CCUACAAACA--AGCUAAACAGGCAAGAAUAGUUUAUAGCGAUGGCAACCAUGGCAAGCAAUAGAGAAAUUGGACGGUGCUGGCAAUCCUGCAGCUGCCGCAUCGUCCAUCCGCUGGAAA ((........--.(((....((((.......))))...((.((((...)))).)).))).....((....((((((((((.((((..........))))))))))))))....))))... ( -34.80) >DroYak_CAF1 4185 120 - 1 CCUACAAACAAAAGCUAAACAGGCAAGCAUAGUUUAUAGCGAUGGCAACCAUGGCAAGCAAUAGAGAAACUGGACGGUGCUGGCAACCCUGCAGCUGCUGCAUCGUCCAUCCGCUGGAAA ((...........(((.....))).(((...((((...((.((((...)))).))))))...........((((((((((.((((.(......).))))))))))))))...)))))... ( -34.80) >consensus CCUACAAACA__AGCUAAACAGGCAAGAAUAGUUUAUAGCGAUGGCAACCAUGGCAAGCAAUAGAGAAAUUGGACGGUGCUGGCAAUCCUGCAGCUGCUGCAUCGUCCAUCCGCUGGAAA ((...........(((....((((.......))))...((.((((...)))).)).))).....((....((((((((((.((((..........))))))))))))))....))))... (-33.32 = -33.10 + -0.22)


| Location | 13,686,867 – 13,686,980 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13686867 113 + 22224390 UUAGUUAAAACUUCAAUGUUCUUCUCUUGUUGUGAUCACAGUUAGUUUUGUGUUGUUGUAGUUGCAUGUAUGCAGUUGCAGUUUUAAUUGUAAUUGCUAUUGGUAAUUGU-------UAA .((((((((((..(((..(......((..((((....))))..))....)..))).((((..((((....))))..))))))))))))))(((((((.....))))))).-------... ( -23.90) >DroSim_CAF1 4063 111 + 1 UUAGUUAAAACUUCAAUGUUCUUUUCUUGUUGUGAUCACAGUUAGUUUUGUGUUGUUGUAGUUGCAU--UUGCAGUUGCAGUUUUAAUUGUAAUUGCUAUUGGUAAUUGU-------UAA .((((((((((..(((..(......((..((((....))))..))....)..))).((((..(((..--..)))..))))))))))))))(((((((.....))))))).-------... ( -23.30) >DroYak_CAF1 4305 119 + 1 UUAGUUAAAACUUCAAUGUUGCUUUCUUGUUGUGAUCACAGUUAGUUUUGUGUUGUUGUAGUUGCA-GUUUGCAGUUGCAGUUUAAAUUGUAAUUGCUAUUGGUAAUUGUUAAUUGUUAA .(((((((....((((((((((...((....(..(.(((((......))))))..)...))..)))-)...(((((((((((....)))))))))))))))))......))))))).... ( -25.90) >consensus UUAGUUAAAACUUCAAUGUUCUUUUCUUGUUGUGAUCACAGUUAGUUUUGUGUUGUUGUAGUUGCAUGUUUGCAGUUGCAGUUUUAAUUGUAAUUGCUAUUGGUAAUUGU_______UAA .((((((((((..(((..(......((..((((....))))..))....)..))).((((..((((....))))..))))))))))))))(((((((.....)))))))........... (-23.00 = -23.33 + 0.33)

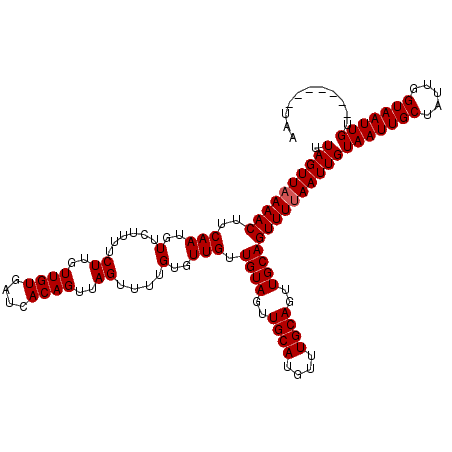

| Location | 13,686,867 – 13,686,980 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -11.30 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13686867 113 - 22224390 UUA-------ACAAUUACCAAUAGCAAUUACAAUUAAAACUGCAACUGCAUACAUGCAACUACAACAACACAAAACUAACUGUGAUCACAACAAGAGAAGAACAUUGAAGUUUUAACUAA ...-------.......................((((((((.(((.((((....))))..........((((........))))....................))).)))))))).... ( -12.70) >DroSim_CAF1 4063 111 - 1 UUA-------ACAAUUACCAAUAGCAAUUACAAUUAAAACUGCAACUGCAA--AUGCAACUACAACAACACAAAACUAACUGUGAUCACAACAAGAAAAGAACAUUGAAGUUUUAACUAA ...-------.......................((((((((.(((.(((..--..)))..........((((........))))....................))).)))))))).... ( -12.10) >DroYak_CAF1 4305 119 - 1 UUAACAAUUAACAAUUACCAAUAGCAAUUACAAUUUAAACUGCAACUGCAAAC-UGCAACUACAACAACACAAAACUAACUGUGAUCACAACAAGAAAGCAACAUUGAAGUUUUAACUAA .......................(((.((........)).)))((((.(((..-(((...........((((........)))).((.......))..)))...))).))))........ ( -9.10) >consensus UUA_______ACAAUUACCAAUAGCAAUUACAAUUAAAACUGCAACUGCAAACAUGCAACUACAACAACACAAAACUAACUGUGAUCACAACAAGAAAAGAACAUUGAAGUUUUAACUAA .................................((((((((.(((.((((....))))..........((((........))))....................))).)))))))).... (-11.53 = -11.87 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:13 2006