
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,685,864 – 13,685,964 |
| Length | 100 |
| Max. P | 0.980379 |

| Location | 13,685,864 – 13,685,964 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -17.15 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13685864 100 + 22224390 AUAAAUAAAUUACAAAAAAUAAUCGAAAAUCGGGGAUCCCACAUCAUGCUCGAU---------------AU-----AUACGAUAUACGAUGCAUGUGGAUCCCCUUUGGAUGGUAGAUCU .........((((........(((.(((...((((((((.....((((((((((---------------((-----.....)))).))).))))).))))))))))).))).)))).... ( -28.10) >DroSim_CAF1 3070 103 + 1 AUAAAUAAAUUACAAAAAAUAAUCGAAAAUCGGGGACCCC--AUCAUGAUCGAU---------------AUACGAUAUACUAUAUACGAUACAUGUGGAUCCCCUUUGGAUGGUAGAUCU .........((((........(((.(((...(((((..((--(.((((((((((---------------(((........))))).)))).))))))).)))))))).))).)))).... ( -24.80) >DroYak_CAF1 3241 118 + 1 AUAAAUAAAUUACAAAAAAUAAUCGAAAAUCGGGGAUCCC--AUCAUAUACGAUAAUGCGAUAUACAAUAUACGAUAUACGAUAUACGAUACGUGUGGAUCCCCUUUGGAUGGUAGAUCU .........((((........(((.(((...((((((((.--(((.(((((((((.((.......)).))).)).)))).)))(((((...)))))))))))))))).))).)))).... ( -24.40) >consensus AUAAAUAAAUUACAAAAAAUAAUCGAAAAUCGGGGAUCCC__AUCAUGAUCGAU_______________AUACGAUAUACGAUAUACGAUACAUGUGGAUCCCCUUUGGAUGGUAGAUCU .........((((........(((.(((...((((((((.....((((((((..................................))).))))).))))))))))).))).)))).... (-17.15 = -18.05 + 0.89)

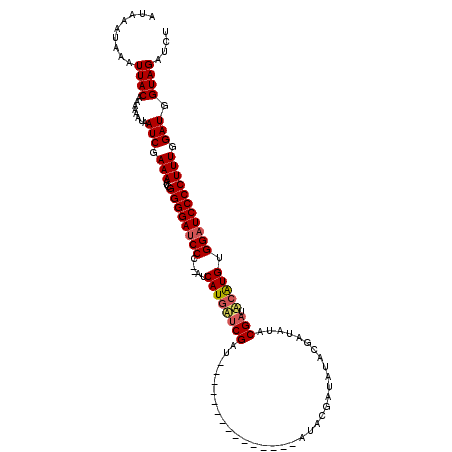

| Location | 13,685,864 – 13,685,964 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13685864 100 - 22224390 AGAUCUACCAUCCAAAGGGGAUCCACAUGCAUCGUAUAUCGUAU-----AU---------------AUCGAGCAUGAUGUGGGAUCCCCGAUUUUCGAUUAUUUUUUGUAAUUUAUUUAU ................((((((((.(((((.((((((((...))-----))---------------).)))))))).....)))))))).......((((((.....))))))....... ( -28.00) >DroSim_CAF1 3070 103 - 1 AGAUCUACCAUCCAAAGGGGAUCCACAUGUAUCGUAUAUAGUAUAUCGUAU---------------AUCGAUCAUGAU--GGGGUCCCCGAUUUUCGAUUAUUUUUUGUAAUUUAUUUAU ................((((((((.((((.(((((((((........))))---------------).))))))))..--.)))))))).......((((((.....))))))....... ( -27.90) >DroYak_CAF1 3241 118 - 1 AGAUCUACCAUCCAAAGGGGAUCCACACGUAUCGUAUAUCGUAUAUCGUAUAUUGUAUAUCGCAUUAUCGUAUAUGAU--GGGAUCCCCGAUUUUCGAUUAUUUUUUGUAAUUUAUUUAU ................((((((((..(((...))).(((((((((.((.(((.(((.....))).)))))))))))))--))))))))).......((((((.....))))))....... ( -30.10) >consensus AGAUCUACCAUCCAAAGGGGAUCCACAUGUAUCGUAUAUCGUAUAUCGUAU_______________AUCGAACAUGAU__GGGAUCCCCGAUUUUCGAUUAUUUUUUGUAAUUUAUUUAU ................((((((((.(((((.(((..................................)))))))).....)))))))).......((((((.....))))))....... (-19.07 = -19.41 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:10 2006