
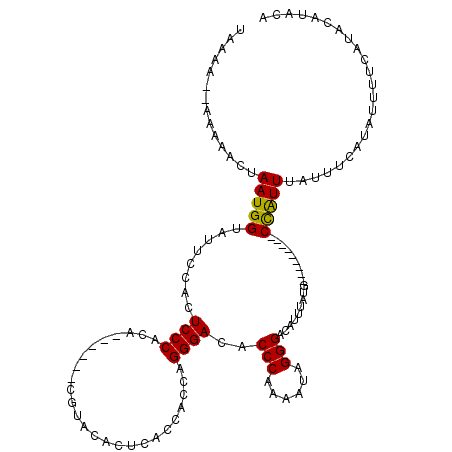
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,683,867 – 13,684,086 |
| Length | 219 |
| Max. P | 0.988082 |

| Location | 13,683,867 – 13,683,970 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -15.53 |
| Consensus MFE | -9.98 |
| Energy contribution | -9.54 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13683867 103 - 22224390 UAAAA--AAAAACUAAUGGUAUUCCACUCCCACA------CGUACACUCACCACCAGGGACACCCAAAAUAGGGAUAUUUAUU---------CCAUUUAUUUCAUAUUUUCAUACAUACA .....--..(((..(((((........((((...------.((......)).....(((...)))......))))........---------)))))..))).................. ( -11.69) >DroSim_CAF1 1150 97 - 1 --------AAAACUAAUGGUAUUCCACUCCCACA------CGUACACUCACCACCAGGGACACCCAAAAUAGGGACAUUUAUG---------CCAUUUGUUUCAUAUUUCCAAACAUACA --------.((((.((((((((.....((((...------.((......)).....(((...)))......)))).....)))---------))))).)))).................. ( -18.90) >DroYak_CAF1 1160 118 - 1 UAAAAACAAAAACUAAUGGUAUUCCACUCCCACAUUGAUACACUCACUCACCACCAGGGACACCCAAAAUAGGGACAUUAGUGCAUUUACCUCUGUUUACUUCAUAUUUUCAUACAUG-- .........((((....((((...(((((((....(((.........)))......))))..(((......)))......)))....))))...))))....................-- ( -16.00) >consensus UAAAA__AAAAACUAAUGGUAUUCCACUCCCACA______CGUACACUCACCACCAGGGACACCCAAAAUAGGGACAUUUAUG_________CCAUUUAUUUCAUAUUUUCAUACAUACA ..............(((((........((((.........................))))..(((......)))..................)))))....................... ( -9.98 = -9.54 + -0.44)

| Location | 13,683,938 – 13,684,050 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -12.47 |
| Consensus MFE | -11.09 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13683938 112 - 22224390 AAUCAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUAUAUAUUCUAGCAUAUAUAUAAAA--AAAAACUAAUGGUAUUCCACUCCCACA------ .........((((((.....))))))...(((((((....)).................((((((((......))))))))....--...........)))))...........------ ( -13.40) >DroSim_CAF1 1221 101 - 1 ---UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGAAAGCAACUCAAGCAAACAAAAAUA--UAUUCUAGCAUAUAUA--------AAAACUAAUGGUAUUCCACUCCCACA------ ---..((((((....(((((..(((((.......((....))...)))))..)))))...))--))))............--------........(((....)))........------ ( -12.00) >DroYak_CAF1 1238 115 - 1 ---UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUA--UAUUCUAGCAUAUAUGUAAAAACAAAAACUAAUGGUAUUCCACUCCCACAUUGAUA ---......((((((.....))))))........((....))...((((..........(((--(((........))))))...............(((....)))........)))).. ( -12.00) >consensus ___UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUA__UAUUCUAGCAUAUAUAUAAAA__AAAAACUAAUGGUAUUCCACUCCCACA______ ......(((((.((((((.((....)).......((....))...........................)))))).)))))...............(((....))).............. (-11.09 = -11.20 + 0.11)



| Location | 13,683,970 – 13,684,086 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -9.67 |
| Consensus MFE | -9.20 |
| Energy contribution | -9.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13683970 116 - 22224390 AACAAAAC----CUAAAAACCCCCAAAAAGCCCAAAAAAAAAUCAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUAUAUAUUCUAGCAUAUAUA ........----.................((..................((((((.....))))))........((....)).............................))....... ( -9.20) >DroSim_CAF1 1247 110 - 1 AACAAAAC----CCAAAAACCC-CGAAAAGCCCAAAAAAA---UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGAAAGCAACUCAAGCAAACAAAAAUA--UAUUCUAGCAUAUAUA ........----..........-.................---..((((((....(((((..(((((.......((....))...)))))..)))))...))--))))............ ( -10.40) >DroYak_CAF1 1278 114 - 1 AACAAGAAAAAAACCAGAACCC-ACAAAAGCCCAAAAAAA---UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUA--UAUUCUAGCAUAUAUG ....((((..............-.................---......((((((.....))))))........((....))....................--..)))).......... ( -9.40) >consensus AACAAAAC____CCAAAAACCC_CAAAAAGCCCAAAAAAA___UAAAUAUAAAUGUUGUUCAUUUGACUAAUACGCGGAAGCAACUCAACCAAACAAAAAUA__UAUUCUAGCAUAUAUA .............................((..................((((((.....))))))........((....)).............................))....... ( -9.20 = -9.20 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:09 2006