
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,678,830 – 13,679,039 |
| Length | 209 |
| Max. P | 0.724442 |

| Location | 13,678,830 – 13,678,927 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -9.43 |
| Energy contribution | -14.67 |
| Covariance contribution | 5.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
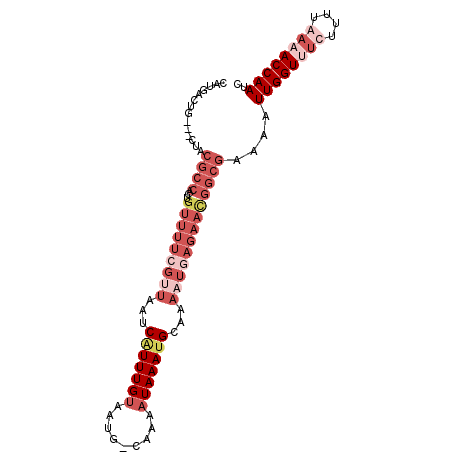
>X_DroMel_CAF1 13678830 97 + 22224390 CAUGACUGCCACUACGCCACUGUUUUCGUUAAUCAUUUGUAAUG-CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUGAAACCAAUC ..............((((...(((((((((...(((((((....-....)))))))...)))))))))))))...(((((((((....))))))))). ( -28.30) >DroSec_CAF1 10621 95 + 1 CAUGACUGC--CUACGCCACUGUUUUCGUUAAUCAUUUGUAAUG-CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUGAAACCAAUC .........--...((((...(((((((((...(((((((....-....)))))))...)))))))))))))...(((((((((....))))))))). ( -28.30) >DroEre_CAF1 9621 73 + 1 -----------CUAGACCAAUCAUUUAGUCAAUCAUUUGCAAUG-CAGAAUAAACGC-------------CGAAAAUUGGUUUCUCUUAAUACCAAGC -----------..((((((((..((..((......(((((...)-))))......))-------------..)).))))))))............... ( -9.60) >DroYak_CAF1 10380 95 + 1 CAUGACCG---CUGCGCCACUGUUUUCGUUAAUCGUUUGUAAUGGCAAAAUAAAUGCAAAUUCAGAAUGGCGAAAAUUGGUUGCUUUUAAUACCAAUC ...(((((---.(.((((((((..((.((......(((((....)))))......)).))..)))..)))))...).)))))................ ( -19.20) >consensus CAUGACUG___CUACGCCACUGUUUUCGUUAAUCAUUUGUAAUG_CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUAAAACCAAUC ..............((((...(((((((((...(((((((.........)))))))...)))))))))))))....((((((((....)))))))).. ( -9.43 = -14.67 + 5.25)

| Location | 13,678,848 – 13,678,964 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.93 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
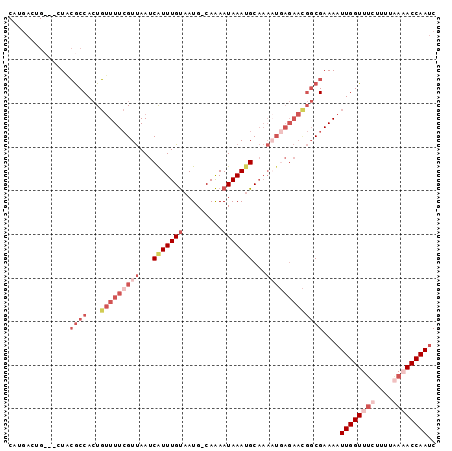
>X_DroMel_CAF1 13678848 116 + 22224390 ACUGUUUUCGUUAAUCAUUUGUAAUG-CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUGAAACCAAUCAAAUUGUUUUG--GCUUCGCAAUAGU-GCAGUAUGCAAUU .(((((((((((...(((((((....-....)))))))...))))))))))).....(((((((((....)))))))))..((((((....--(((.(((....))-).)))..)))))) ( -33.20) >DroSec_CAF1 10637 109 + 1 ACUGUUUUCGUUAAUCAUUUGUAAUG-CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUGAAACCAAUCAAAUUGUU-UG--GCUUCGCAAUAGU-GCAGUAU------ .(((((((((((...(((((((....-....)))))))...))))))))))).....(((((((((....))))))))).........-..--(((.(((....))-).)))..------ ( -29.10) >DroYak_CAF1 10395 118 + 1 ACUGUUUUCGUUAAUCGUUUGUAAUGGCAAAAUAAAUGCAAAUUCAGAAUGGCGAAAAUUGGUUGCUUUUAAUACCAAUCGAAUUGUU-UAAUUCUACGCAAUAGCCACAGCAUGCAG-U (((((....(((.....((((.....(((.......))).....)))).((((....((((((..........))))))...(((((.-((....)).))))).)))).)))..))))-) ( -19.90) >consensus ACUGUUUUCGUUAAUCAUUUGUAAUG_CAAAAUAAAUGCAAAAUGAGAACGGCGAAAAUUGGUUUCUUUUGAAACCAAUCAAAUUGUU_UG__GCUUCGCAAUAGU_GCAGUAUGCA__U .(((((((((((...(((((((.........)))))))...))))))))))).....(((((((((....)))))))))...(((((...........)))))................. (-18.71 = -19.93 + 1.23)


| Location | 13,678,927 – 13,679,039 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -9.57 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
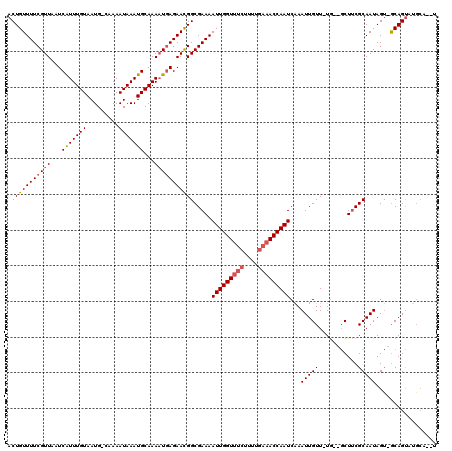
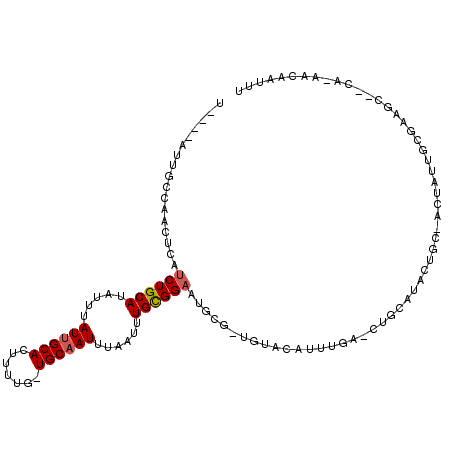
>X_DroMel_CAF1 13678927 112 - 22224390 UAUAUAUUGCCAACUCAUCUGCAUAUUUAUUGCAUUUUGUGUGCAAUUUAAUUUGUGGCAUGCAGUGUACAUUUAAAUUGCAUACUGC-ACUAUUGCGAAGC--CAAAACAAUUU ......((((........(..((.(((.(((((((.....)))))))..))).))..)..((((((((.((.......)).)))))))-).....))))...--........... ( -24.70) >DroSec_CAF1 10716 92 - 1 UAUAUAUUGCCGACUCAUCUGCAUAUUUAUUGCACUUUUG-UGCAAUUUAAUUUGUGGAAUGC------------------AUACUGC-ACUAUUGCGAAGC--CA-AACAAUUU ......((((.......((..((.(((.(((((((....)-))))))..))).))..)).(((------------------.....))-).....))))...--..-........ ( -20.40) >DroEre_CAF1 9694 87 - 1 U----AUUGCGACCUCAUCUGCAUAUUUAUUGCACUUUUG-UGCCAUUUAAUUUGCGGAAUUCGCUGUACAAUUGA-CUAC----UG-----------------UA-AACAAUUU (----((.((((.....((((((.(((.((.((((....)-))).))..))).))))))..)))).))).......-....----..-----------------..-........ ( -18.00) >DroYak_CAF1 10475 108 - 1 U----AUUGCAAACUCAUCUGCAUAUUUAUUGCACUUUUG-UGCAAUUUCAUUUGCGGAAUUCGAUGAACAUUUGA-CUGCAUGCUGUGGCUAUUGCGUAGAAUUA-AACAAUUC .----.((((.......((((((.....(((((((....)-))))))......))))))..((((.......))))-..(((.((....))...))))))).....-........ ( -24.10) >consensus U____AUUGCCAACUCAUCUGCAUAUUUAUUGCACUUUUG_UGCAAUUUAAUUUGCGGAAUGCG_UGUACAUUUGA_CUGCAUACUGC_ACUAUUGCGAAGC__CA_AACAAUUU .................((((((.....((((((.......))))))......))))))........................................................ ( -9.57 = -9.83 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:05 2006