
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,668,362 – 13,668,471 |
| Length | 109 |
| Max. P | 0.737680 |

| Location | 13,668,362 – 13,668,471 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -12.62 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
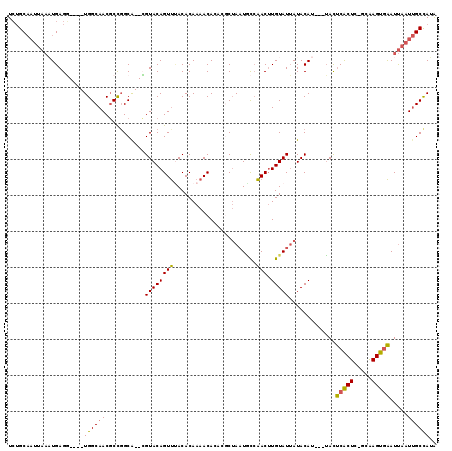
>X_DroMel_CAF1 13668362 109 + 22224390 UCUGCAAUUAAAUGAGU----UGGCAACGCCGGCA--AGUACAGUUCACACAAAACACCCGCUAAUACGAACUUGUAUUAUACAU---UAUUCACUC-GCAAGUGAAUUAAUUGCCAUA ...(((((((((((.((----((((...)))))).--((((((((((.....................)))).))))))...)))---)(((((((.-...)))))))))))))).... ( -26.60) >DroSec_CAF1 3498 105 + 1 UCUGCAAUUAAAUGAGG----UGGCAACGCCGGCA--CGUACAGUUUACACAAAACA----CUAAUGCCAACUUGUAUUAUACAU---UAUUCACUC-GCAAGUGAAUUAAUUGCCAUA ...(((((((((((.((----((....))))((((--......((((.....)))).----....)))).............)))---)(((((((.-...)))))))))))))).... ( -27.50) >DroSim_CAF1 4145 105 + 1 UCUGCAAUUAAAUGAGG----UGGCAACGCCGGCA--CGUACAGUUUACACAAAACA----CUAAUGCCAACUUGUAUUAUACAU---UAUUCACUC-GCAAGUGAAUUAAUUGCCAUA ...(((((((((((.((----((....))))((((--......((((.....)))).----....)))).............)))---)(((((((.-...)))))))))))))).... ( -27.50) >DroEre_CAF1 3569 109 + 1 UCUGCAAUUAAUUGAGG----UGGCAACGCUUGCG--CGUACAGUUUACACAAAACACACGCUAAUGCCAACUUGUAUUAUACAU---UACUCACUC-GCAAGUGAAUUAAUUGCCAAA ...((((((((((....----.(....)(((((((--(((...((((.....))))..))))((((((......)))))).....---.........-)))))).)))))))))).... ( -27.10) >DroYak_CAF1 3484 112 + 1 UCUGCAAUUAAAUGAGG----UGGCAACGCCGGCG--CGUACAGUUUACACAAAACACACGCUAAUACCAACUUGUAUUAUACAUUAUUACUCACUC-GCAAGUGAAUUAAUUGCUAUG ...(((((((((((.((----((....))))...(--(((...((((.....))))..))))((((((......))))))..)))).....(((((.-...)))))..))))))).... ( -28.00) >DroAna_CAF1 6637 109 + 1 UGAGC-------UGAGCUGCCUGGCAACUCUGGCGGAGGUACAGUCGACACAAAACACAGGCAACUCUCGACAUGUAUGAUACAG---GACCCGCUCCGGGAGCAGAUCGGUGGCUAGA ...((-------(((.(((((((....(((.....)))((((((((((..........((....)).))))).)))))....)))---..((((...)))).)))).)))))....... ( -31.90) >consensus UCUGCAAUUAAAUGAGG____UGGCAACGCCGGCA__CGUACAGUUUACACAAAACACACGCUAAUGCCAACUUGUAUUAUACAU___UACUCACUC_GCAAGUGAAUUAAUUGCCAUA .....................((((((...........((((((((.......................))).))))).............(((((.....))))).....)))))).. (-12.62 = -11.90 + -0.72)


| Location | 13,668,362 – 13,668,471 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -18.13 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
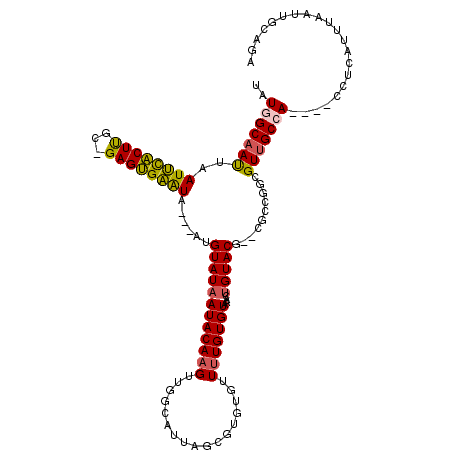
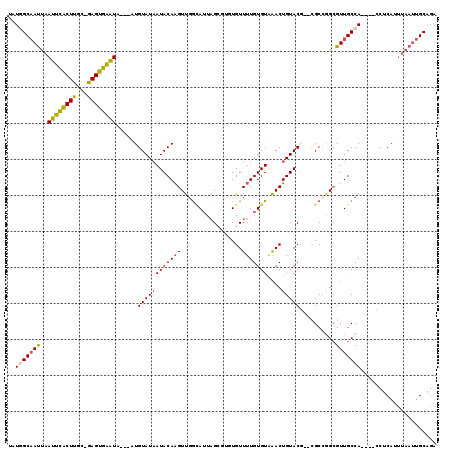
>X_DroMel_CAF1 13668362 109 - 22224390 UAUGGCAAUUAAUUCACUUGC-GAGUGAAUA---AUGUAUAAUACAAGUUCGUAUUAGCGGGUGUUUUGUGUGAACUGUACU--UGCCGGCGUUGCCA----ACUCAUUUAAUUGCAGA ....((((((((((((((...-.))))))((---((((.((((((......))))))(((((((....((....))..))))--)))..))))))...----......))))))))... ( -26.60) >DroSec_CAF1 3498 105 - 1 UAUGGCAAUUAAUUCACUUGC-GAGUGAAUA---AUGUAUAAUACAAGUUGGCAUUAG----UGUUUUGUGUAAACUGUACG--UGCCGGCGUUGCCA----CCUCAUUUAAUUGCAGA ..(((((((..(((((((...-.))))))).---.............((((((((...----.((((.....)))).....)--))))))))))))))----................. ( -27.80) >DroSim_CAF1 4145 105 - 1 UAUGGCAAUUAAUUCACUUGC-GAGUGAAUA---AUGUAUAAUACAAGUUGGCAUUAG----UGUUUUGUGUAAACUGUACG--UGCCGGCGUUGCCA----CCUCAUUUAAUUGCAGA ..(((((((..(((((((...-.))))))).---.............((((((((...----.((((.....)))).....)--))))))))))))))----................. ( -27.80) >DroEre_CAF1 3569 109 - 1 UUUGGCAAUUAAUUCACUUGC-GAGUGAGUA---AUGUAUAAUACAAGUUGGCAUUAGCGUGUGUUUUGUGUAAACUGUACG--CGCAAGCGUUGCCA----CCUCAAUUAAUUGCAGA ....((((((((((((((...-.))))))).---............((.(((((...((((((((...((....))...)))--)))..))..)))))----.))....)))))))... ( -31.40) >DroYak_CAF1 3484 112 - 1 CAUAGCAAUUAAUUCACUUGC-GAGUGAGUAAUAAUGUAUAAUACAAGUUGGUAUUAGCGUGUGUUUUGUGUAAACUGUACG--CGCCGGCGUUGCCA----CCUCAUUUAAUUGCAGA ....((((((((((((((...-.)))))))................((.(((((...((((((((...((....))...)))--)))..))..)))))----.))....)))))))... ( -29.60) >DroAna_CAF1 6637 109 - 1 UCUAGCCACCGAUCUGCUCCCGGAGCGGGUC---CUGUAUCAUACAUGUCGAGAGUUGCCUGUGUUUUGUGUCGACUGUACCUCCGCCAGAGUUGCCAGGCAGCUCA-------GCUCA ...(((....(((((((((...)))))))))---..................((((((((((.((((((.((.((.......)).))))))...)))))))))))).-------))).. ( -35.70) >consensus UAUGGCAAUUAAUUCACUUGC_GAGUGAAUA___AUGUAUAAUACAAGUUGGCAUUAGCGUGUGUUUUGUGUAAACUGUACG__CGCCGGCGUUGCCA____CCUCAUUUAAUUGCAGA ..(((((((..((((((((...))))))))......((((((((((((.................)))))))....)))))..........)))))))..................... (-18.13 = -17.75 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:59 2006