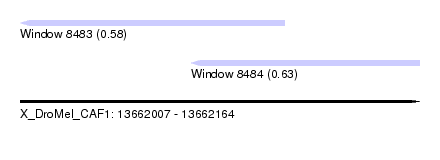
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,662,007 – 13,662,164 |
| Length | 157 |
| Max. P | 0.628978 |

| Location | 13,662,007 – 13,662,111 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.65 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13662007 104 - 22224390 GUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGGGGCGACAAACUUUGGUCAAACGGUUGCCGCUAACCGGUUUAAUUUUAAGUCCUUUACCUGAACCGAU ..........((((........(((((((((((((..((((((....(((....)))..)))))))))))))))))))..((((.((.....)).)))).)))) ( -28.80) >DroSec_CAF1 4987 102 - 1 GUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGGGGCGACAAACUUUGGUCAAACGGUU--UGGUAACCGGUUUAAUUUUAAGUCCUUUACCUGAACCGAU ...........((((.(((((.......)))))))))...(((........)))...(((((--((((((..(..(((....)))..)..))))).)))))).. ( -25.40) >DroEre_CAF1 5112 104 - 1 GUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGGGGCGACAAACUUUGGUCAAACGGUUGCCGCUAACCGGUUUAUUUUCCAGUCCUUUACCUGAACCGAU ..........((((.......((((((((((((((..((((((....(((....)))..))))))))))))))))))))....(((........)))...)))) ( -31.30) >DroYak_CAF1 4882 104 - 1 GUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGGGGCGACAAACUUUGGUCAAACGGUUGCCGCUAACCGGUUUAUUUUCAAGUCCUUUACCUGAACCGAU ..........((((.......((((((((((((((..((((((....(((....)))..)))))))))))))))))))).((((.((.....)).)))).)))) ( -31.40) >consensus GUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGGGGCGACAAACUUUGGUCAAACGGUUGCCGCUAACCGGUUUAAUUUCAAGUCCUUUACCUGAACCGAU ..........((((........(((((((((((((((((((((........))).....))).)))))))))))))))..((((.((.....)).)))).)))) (-25.34 = -25.65 + 0.31)


| Location | 13,662,074 – 13,662,164 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -26.12 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13662074 90 - 22224390 AGAGGUAUGACG-----GGCUGAGCUUGGUUUCUCCAAAGGGGUUGGGUUC-------CGUUCGUGUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGG .(((..((((((-----(((.(((((..(..(((....)))..)..)))))-------.))))..)))))..)))..(((.(((((.......)))))))). ( -32.70) >DroSec_CAF1 5052 89 - 1 AGAGGUAUGACG-----GGCUGAGCUUGGUUUCUCCA-AGGGGUUGGGUUC-------CGUUCGUGUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGG .(((..((((((-----(((.(((((..(..((....-.))..)..)))))-------.))))..)))))..)))..(((.(((((.......)))))))). ( -32.10) >DroEre_CAF1 5179 89 - 1 AGAGGUAUGAUG-----GGUUGAGCUUGGUUUCUCCA-AGGGGUUGGGUUC-------CGUUCGUGUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGG .(((..((((((-----..(.(((((..(..((....-.))..)..)))))-------.)..)..)))))..)))..(((.(((((.......)))))))). ( -28.70) >DroYak_CAF1 4949 101 - 1 AGAGGUAUGAUGGGACGGGUUGAGCUUGGUUUCUCCA-AGGGGUUGGGUUCCACCAACCGUUCGUGUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGG .(((...((((((.(((..(....(((((.....)))-)).((((((......)))))))..))).)))))))))..(((.(((((.......)))))))). ( -39.80) >consensus AGAGGUAUGACG_____GGCUGAGCUUGGUUUCUCCA_AGGGGUUGGGUUC_______CGUUCGUGUCAUCACUCAUCGCUCAAUCAUAAACCGGUUGGCGG .(((..((((((.........(((((..(..(((....)))..)..))))).............))))))..)))..(((.(((((.......)))))))). (-26.12 = -25.88 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:57 2006