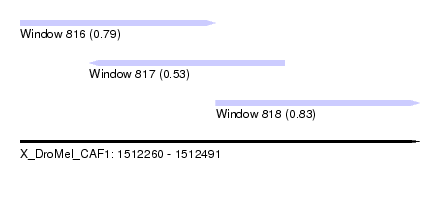
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,512,260 – 1,512,491 |
| Length | 231 |
| Max. P | 0.832077 |

| Location | 1,512,260 – 1,512,373 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1512260 113 + 22224390 CUCUCUACUGAGGCCAGCCGUCCCUAUUCGGCUGAAUCUGCAUCUGUCUGUACAAGCAUUUCUUCUUCCUCUGUAUCUAU-------CCUUUAUCUCCUUGCCAAACACACUAACGAUCC .....(((.((((.((((((........))))))....(((...(((....))).))).........)))).))).....-------................................. ( -18.60) >DroSec_CAF1 1766 113 + 1 CUCUCUACUGAGGCCAGCCGUCCCUACUCGGCUGAAUCUGCAUCUGUCUGUACAAGCAUUUCUUCUUCCUCUGUAUCUAU-------CCUUUAUCUCCUUGCCAAACACACUAACGAUCC .....(((.((((.((((((........))))))....(((...(((....))).))).........)))).))).....-------................................. ( -18.60) >DroEre_CAF1 1761 113 + 1 CUCUCUACUGAGCCCAGCCGUCUCUACUCGGCUGUAUCUGCAUCUGUCUGUACAAGCAUUUCUUCUUCCUCUGUACCUAU-------CCUUUAUCUCCUUGCCAAUCACACUAACGAUCC .....(((.(((..((((((........))))))....(((...(((....))).)))..........))).))).....-------................................. ( -14.90) >DroYak_CAF1 3258 120 + 1 CUCUCUACUGAGCCCAGCCGUUUCUACUCGGCUGUAUCUGCAUCUGUCUGUACAAGCAUUUCUUCUUCCUCUGUAUCUAUAUCUGUACCUUUAUCUCCCUGCCAAUCACACUAACGAUCC ........((.((.((((((........))))))....((((..(((..(((((((.(........).)).)))))..)))..)))).............))))................ ( -16.00) >consensus CUCUCUACUGAGCCCAGCCGUCCCUACUCGGCUGAAUCUGCAUCUGUCUGUACAAGCAUUUCUUCUUCCUCUGUAUCUAU_______CCUUUAUCUCCUUGCCAAACACACUAACGAUCC .....(((.((((.((((((........))))))....(((...(((....))).))).........)))).)))............................................. (-15.68 = -16.18 + 0.50)

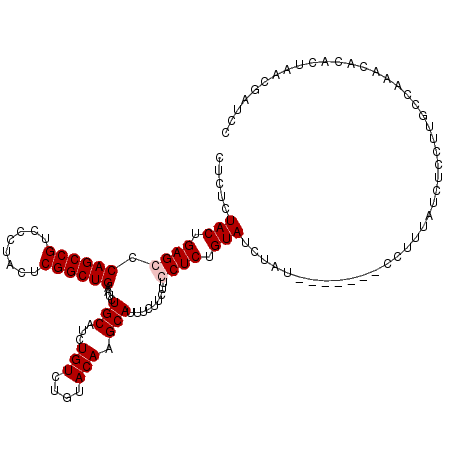
| Location | 1,512,300 – 1,512,413 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -22.56 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1512300 113 - 22224390 AGAUUAGAGCGUUUCGCGAAGAAGGCGAAGCACGCUUAAUGGAUCGUUAGUGUGUUUGGCAAGGAGAUAAAGG-------AUAGAUACAGAGGAAGAAGAAAUGCUUGUACAGACAGAUG ......(((((((((.........((.(((((((((.((((...))))))))))))).))...........(.-------.......)..........)))))))))............. ( -26.20) >DroSec_CAF1 1806 113 - 1 AGAUUAGAGCGUUUCGCGAAGAAGGCGAAGCACGCUUAAUGGAUCGUUAGUGUGUUUGGCAAGGAGAUAAAGG-------AUAGAUACAGAGGAAGAAGAAAUGCUUGUACAGACAGAUG ......(((((((((.........((.(((((((((.((((...))))))))))))).))...........(.-------.......)..........)))))))))............. ( -26.20) >DroEre_CAF1 1801 113 - 1 AGAUUAGAGCGUAUCGCGAAGAAGGCGAAGCACGCUUAAUGGAUCGUUAGUGUGAUUGGCAAGGAGAUAAAGG-------AUAGGUACAGAGGAAGAAGAAAUGCUUGUACAGACAGAUG .((((.((((((.((((.......))))...))))))....))))((((((...)))))).............-------....(((((.((.(........).)))))))......... ( -20.50) >DroYak_CAF1 3298 120 - 1 AGAUUAGAGCGUGUCGCGAAGAAGGCGAAGCACGCUGGAUGGAUCGUUAGUGUGAUUGGCAGGGAGAUAAAGGUACAGAUAUAGAUACAGAGGAAGAAGAAAUGCUUGUACAGACAGAUG .((((..((((((((((.......)))..))))))).....))))(((..((((...((((...........((((.......).)))..............))))..)))))))..... ( -22.31) >consensus AGAUUAGAGCGUUUCGCGAAGAAGGCGAAGCACGCUUAAUGGAUCGUUAGUGUGAUUGGCAAGGAGAUAAAGG_______AUAGAUACAGAGGAAGAAGAAAUGCUUGUACAGACAGAUG ......(((((((((.(......)((.(((((((((.((((...))))))))))))).))......................................)))))))))............. (-22.56 = -23.38 + 0.81)

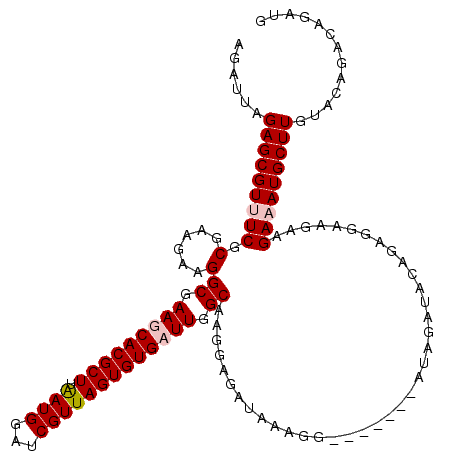
| Location | 1,512,373 – 1,512,491 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1512373 118 + 22224390 AUUAAGCGUGCUUCGCCUUCUUCGCGAAACGCUCUAAUCUGUACAAAUUGUUUCUAGAUGAAAACAAUUAACCCGACCAACCACUGCGCAACAUC--AGUCAGUAUAUGGUCAGCUUGGC .(((((((((.(((((.......))))).))).............(((((((((.....).)))))))).....(((((...((((.((......--.))))))...))))).)))))). ( -28.80) >DroSec_CAF1 1879 118 + 1 AUUAAGCGUGCUUCGCCUUCUUCGCGAAACGCUCUAAUCUGUACACAUUGUUUCUAUACAAGAACAAUUAACCCGACCAACCACUGCGCAAAAUC--AGUCAGUAUAUGGUCAGCUUGGC .(((((((((.(((((.......))))).)))..............(((((((........)))))))......(((((...((((.((......--.))))))...))))).)))))). ( -27.30) >DroEre_CAF1 1874 118 + 1 AUUAAGCGUGCUUCGCCUUCUUCGCGAUACGCUCUAAUCUGUACAAAUUGUUUCUAGAUGAAAACAAUUAACCCGACCAGCCACUGUGCAAAAUC--AGUCAGUGUAUGGUUAGCUUGGC .(((((((((..((((.......))))..))).............(((((((((.....).)))))))).....(((((..(((((.((......--.)))))))..))))).)))))). ( -30.40) >DroYak_CAF1 3378 120 + 1 AUCCAGCGUGCUUCGCCUUCUUCGCGACACGCUCUAAUCUGUACAAAUUGUUUCUAGAUGAAAACAAUUAACCCGGCCAGCCACUGUGCAAAAUCACAGUCAGUAUAUGGUCAGCUUGGC ....((((((..((((.......))))))))))((((.((.....(((((((((.....).)))))))).....(((((((.((((((......))))))..))...))))))).)))). ( -30.40) >consensus AUUAAGCGUGCUUCGCCUUCUUCGCGAAACGCUCUAAUCUGUACAAAUUGUUUCUAGAUGAAAACAAUUAACCCGACCAACCACUGCGCAAAAUC__AGUCAGUAUAUGGUCAGCUUGGC .(((((((((.(((((.......))))).))).............((((((((........)))))))).....(((((...((((.((.........))))))...))))).)))))). (-26.35 = -26.60 + 0.25)

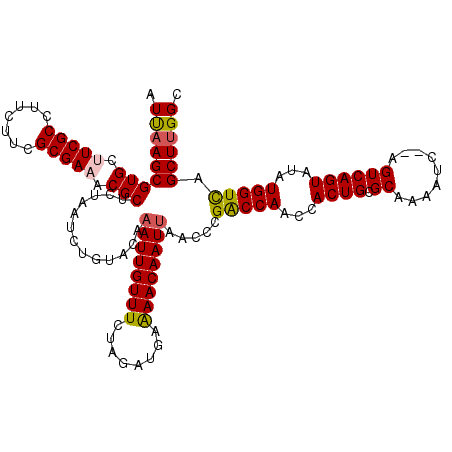
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:46 2006