
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,619,768 – 13,619,953 |
| Length | 185 |
| Max. P | 0.999936 |

| Location | 13,619,768 – 13,619,888 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13619768 120 + 22224390 AAACAACAAACAUCAAAAGUUGCCAACUUUUUUCUCUUGCGACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGC ..................((((......((((((...(((.......)))...))))))(((((((((((......)))).))).)))).(((((((...)))))))..))))....... ( -39.80) >DroSec_CAF1 5639 120 + 1 AAGCAACAAACAUCAAAAGUUGCCAACUUUUUUCUGUCGCGACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGC ..(((((...........))))).....(((((((((.((....)).)))...))))))(((((((((((......)))).))).)))).(((((((...)))))))............. ( -41.10) >DroSim_CAF1 5635 120 + 1 AAACAACAAACAUCAAAAGUUGCCAACUUUUUUCUCUUGCGACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGC ..................((((......((((((...(((.......)))...))))))(((((((((((......)))).))).)))).(((((((...)))))))..))))....... ( -39.80) >DroEre_CAF1 5640 120 + 1 AAACAACAAACAUCAAAAGUUGCCAACUUUUUUCUCUUGCGACCGCAGCAGCCGAAGAAGCAGCGGCGUCAGCAGCGACGGCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGC ..................((((......((((((..((((.......))))..))))))(((((((((((......))).)))).)))).(((((((...)))))))..))))....... ( -40.60) >DroYak_CAF1 5347 120 + 1 AAACAACAAACAUCAAAAGUUGCCAACUUUUUUCUCUUGCGACCGCAGCAGCCAAAGGAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGACGCGCUGGCAACAACAAAUUGC ..................((((....(((((..((.((((....)))).))..))))).(((((((((((......)))).))).)))).(((((((...)))))))..))))....... ( -40.50) >consensus AAACAACAAACAUCAAAAGUUGCCAACUUUUUUCUCUUGCGACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGC ..................((((......((((((...(((.......)))...))))))(((((((((((......)))).))).)))).(((((((...)))))))..))))....... (-38.08 = -38.32 + 0.24)

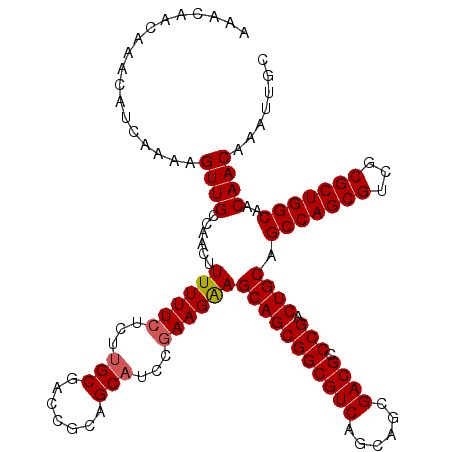

| Location | 13,619,768 – 13,619,888 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -48.60 |
| Energy contribution | -48.88 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13619768 120 - 22224390 GCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUCGCAAGAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGUUU (((((.((((((((((((((((((((((((....))).))))))))..((..((((((.((.((....)).)).)))).))..))........)))))))))....))))...))))).. ( -51.20) >DroSec_CAF1 5639 120 - 1 GCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUCGCGACAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGCUU .((((((.((((((((((((((((((((((....))).)))))))).))))..(((((.((.((....)).)).))))).)))))))....))))))(((((...........))))).. ( -56.20) >DroSim_CAF1 5635 120 - 1 GCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUCGCAAGAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGUUU (((((.((((((((((((((((((((((((....))).))))))))..((..((((((.((.((....)).)).)))).))..))........)))))))))....))))...))))).. ( -51.20) >DroEre_CAF1 5640 120 - 1 GCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGCCGUCGCUGCUGACGCCGCUGCUUCUUCGGCUGCUGCGGUCGCAAGAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGUUU (((((.((((((((((((((((((((((((....))))).))))))..((..((((((.((..(....)..)).)))).))..))........)))))))))....))))...))))).. ( -49.60) >DroYak_CAF1 5347 120 - 1 GCAAUUUGUUGUUGCCAGCGCGUCGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUCCUUUGGCUGCUGCGGUCGCAAGAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGUUU (((((.(((((((((((((((((((..((..(((((..(((((((....)))))))))))).))..)))).)).....((....)).......)))))))))....))))...))))).. ( -45.30) >consensus GCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUCGCAAGAGAAAAAAGUUGGCAACUUUUGAUGUUUGUUGUUU (((((.((((((((((((((((((((((((....))))).))))))(((....(((((.((.((....)).)).))))).)))..........)))))))))....))))...))))).. (-48.60 = -48.88 + 0.28)



| Location | 13,619,808 – 13,619,924 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -44.18 |
| Consensus MFE | -42.60 |
| Energy contribution | -42.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13619808 116 + 22224390 GACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCACACUUUUCCAUUGUCUGAA- .(((((((...........(((((((((((......)))).))).)))).(((((((...))))))).........)))))))..---(((.....)))....................- ( -43.60) >DroSec_CAF1 5679 117 + 1 GACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUGGCACACUUUUCCAUUGUCUGAGG ..((.(((((...(((((.(((((((((((......)))).))).)))).(((((((...))))))).....((((.(((.....---)))..))))......)))))...)).))).)) ( -43.90) >DroSim_CAF1 5675 117 + 1 GACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCACACUUUUCCAUUGUCUGGGG .(((((((...........(((((((((((......)))).))).)))).(((((((...))))))).........)))))))..---(((.....))).......((((.....)))). ( -45.70) >DroEre_CAF1 5680 116 + 1 GACCGCAGCAGCCGAAGAAGCAGCGGCGUCAGCAGCGACGGCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCAUAUUUUUCCAUUGUCUAAA- .(((((((...........(((((((((((......))).)))).)))).(((((((...))))))).........)))))))..---(((.....)))....................- ( -43.60) >DroYak_CAF1 5387 119 + 1 GACCGCAGCAGCCAAAGGAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGACGCGCUGGCAACAACAAAUUGCGGUAAGAAGCAUUCGUUGCACUUUUUUCCAUUGUCUGAA- ........(((.(((.((((((((((((((......)))).))).)))).(((((((...)))))))..((((....(((........)))...))))........))).))).)))..- ( -44.10) >consensus GACCGCAGCAUCCGAAGAAGCAGCGGCGUCAGCAGCGACGCCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA___GCAUUUUUUGCACACUUUUCCAUUGUCUGAA_ .(((((((...........(((((((((((......)))).))).)))).(((((((...))))))).........))))))).....(((.....)))..................... (-42.60 = -42.80 + 0.20)


| Location | 13,619,808 – 13,619,924 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -46.74 |
| Consensus MFE | -40.91 |
| Energy contribution | -41.51 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13619808 116 - 22224390 -UUCAGACAAUGGAAAAGUGUGCAAAAAAUGC---UUACCGCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUC -....(((((..((...(((.(((.....)))---....)))..))..)))))..(((((((((((((((....))).)))))))).))))..(((((.((.((....)).)).))))). ( -47.90) >DroSec_CAF1 5679 117 - 1 CCUCAGACAAUGGAAAAGUGUGCCAAAAAUGC---UUACCGCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUC ...........((..((((((.......))))---)).))(((((.....)))))(((((((((((((((....))).)))))))).))))..(((((.((.((....)).)).))))). ( -46.60) >DroSim_CAF1 5675 117 - 1 CCCCAGACAAUGGAAAAGUGUGCAAAAAAUGC---UUACCGCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUC ..(((.....)))....(((.(((.....)))---.))).(((((.....)))))(((((((((((((((....))).)))))))).))))..(((((.((.((....)).)).))))). ( -48.30) >DroEre_CAF1 5680 116 - 1 -UUUAGACAAUGGAAAAAUAUGCAAAAAAUGC---UUACCGCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGCCGUCGCUGCUGACGCCGCUGCUUCUUCGGCUGCUGCGGUC -....(((((..((.......(((.....)))---.........))..)))))..(((((((((((((((....))))).)))))).))))..(((((.((..(....)..)).))))). ( -43.99) >DroYak_CAF1 5387 119 - 1 -UUCAGACAAUGGAAAAAAGUGCAACGAAUGCUUCUUACCGCAAUUUGUUGUUGCCAGCGCGUCGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUCCUUUGGCUGCUGCGGUC -..(((.(((.(((.......(((((((((((........)).))))))))).(((((((...))))))).(((((..(((((((....))))))))))))))).))).)))........ ( -46.90) >consensus _UUCAGACAAUGGAAAAGUGUGCAAAAAAUGC___UUACCGCAAUUUGUUGUUGCCAGCGCGACGCUGGCUGCAGUCGGGCGUCGCUGCUGACGCCGCUGCUUCUUCGGAUGCUGCGGUC .....(((((..((......(((.................))).))..)))))..(((((((((((((((....))))).)))))).))))..(((((.((.((....)).)).))))). (-40.91 = -41.51 + 0.60)



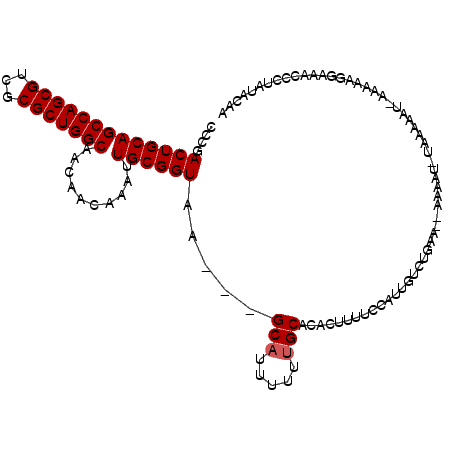
| Location | 13,619,848 – 13,619,953 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13619848 105 + 22224390 CCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCACACUUUUCCAUUGUCUGAA--AAAAU-UAAA----AAAAGGGAAGCCCUAUACAA (((.(((((((((((((...)))))))..........))))))..---(((.....)))....(((((........)))--))...-....----....)))............. ( -25.60) >DroSec_CAF1 5719 104 + 1 CCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUGGCACACUUUUCCAUUGUCUGAGGGAA--------AUUAAAAAAGGAAACCCUAUACAA (((....((((((((((...))))))).....((((.(((.....---)))..))))..............)))....)))..--------........(((....)))...... ( -26.10) >DroSim_CAF1 5715 111 + 1 CCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCACACUUUUCCAUUGUCUGGGGGAAAAU-UAAAAUUAAAAAAGGAAACCCUAUACAA ....(((((((((((((...)))))))..........))))))..---(((.....)))...........((((.(((((......-..................))))).)))) ( -28.26) >DroEre_CAF1 5720 99 + 1 GCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA---GCAUUUUUUGCAUAUUUUUCCAUUGUCUAAA--AAAAC-UAAAAAU---AAUAUAUAA-------AA ((..(((((((((((((...)))))))..........))))))..---)).(((((((.(((.........))).))))--)))..-.......---.........-------.. ( -22.50) >DroYak_CAF1 5427 110 + 1 CCCGACUGCAGCCAGCGACGCGCUGGCAACAACAAAUUGCGGUAAGAAGCAUUCGUUGCACUUUUUUCCAUUGUCUGAA--AAAGUUUUAAAAU---AAGAAAUACCCUAUACAG .....(((..(((((((...))))))).............((((....(((.....)))((((((((.........)))--)))))........---......)))).....))) ( -23.40) >consensus CCCGACUGCAGCCAGCGUCGCGCUGGCAACAACAAAUUGCGGUAA___GCAUUUUUUGCACACUUUUCCAUUGUCUGAA__AAAAU_UAAAAAU_AAAAAGGAAACCCUAUACAA ....(((((((((((((...)))))))..........)))))).....(((.....)))........................................................ (-19.30 = -19.50 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:27 2006