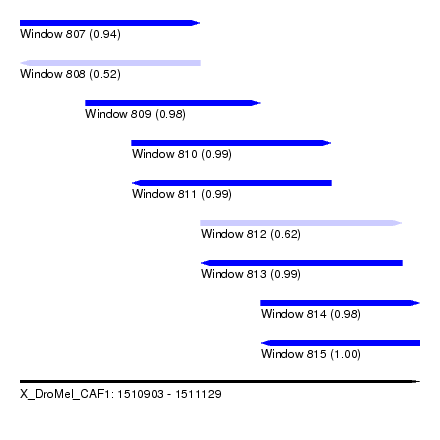
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,510,903 – 1,511,129 |
| Length | 226 |
| Max. P | 0.998461 |

| Location | 1,510,903 – 1,511,005 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1510903 102 + 22224390 AUGA---CCCCGACAACCAUUACCACGCCGACUCAAACUCCAAGCCAGGCUCAGACUCCUACCUCAAUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA ....---...................(((..................))).....................(((((((((((.....))))).))))))...... ( -19.17) >DroSec_CAF1 349 102 + 1 AUGA---CCCCGACCACCAUUACCACGCCGACUCAAACACCAAGCCAGGCUCCGACUCCUACCUCAAUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA ....---...................(((..................))).....................(((((((((((.....))))).))))))...... ( -19.17) >DroSim_CAF1 1923 102 + 1 AUGA---CCCCGACCACCAUUACCACGCCGACUCAAACACCAAGCCAGGCUCAGACUCCUACCUCAAUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA ....---...................(((..................))).....................(((((((((((.....))))).))))))...... ( -19.17) >DroEre_CAF1 361 105 + 1 UUGACCACCCCGACCACCGUAACCACGCCGACUCACACUCCAAGCCAGGCUCAGACUCCUACCUCAAAUCCGAUGGCAGAUCACCAGGAUCUGCCCAUCUACCCA ((((......((.....)).......(((..................))).............))))....(((((((((((.....)))))).)))))...... ( -19.57) >consensus AUGA___CCCCGACCACCAUUACCACGCCGACUCAAACACCAAGCCAGGCUCAGACUCCUACCUCAAUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA ..........................(((..................))).....................(((((((((((.....))))).))))))...... (-17.55 = -17.80 + 0.25)

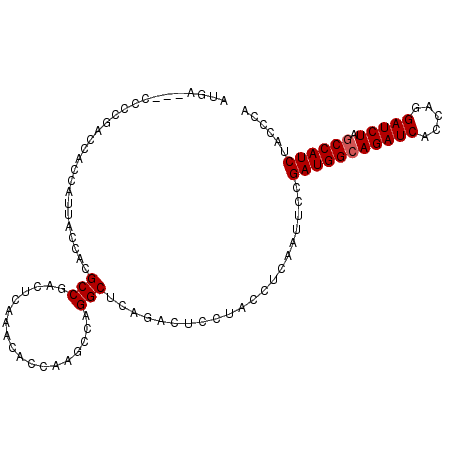
| Location | 1,510,903 – 1,511,005 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -30.29 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1510903 102 - 22224390 UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAUUGAGGUAGGAGUCUGAGCCUGGCUUGGAGUUUGAGUCGGCGUGGUAAUGGUUGUCGGGG---UCAU ..(((.....))).((((((((..(((((((((((((.((.......)).)))))(((.((((..(...)..))))))).)))))..)))..))))))---)).. ( -35.30) >DroSec_CAF1 349 102 - 1 UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAUUGAGGUAGGAGUCGGAGCCUGGCUUGGUGUUUGAGUCGGCGUGGUAAUGGUGGUCGGGG---UCAU ..(((((((.(((((...))))).))))))).(((((((..((.((((........)))).))..)).))))).(((((............)))))..---.... ( -32.60) >DroSim_CAF1 1923 102 - 1 UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAUUGAGGUAGGAGUCUGAGCCUGGCUUGGUGUUUGAGUCGGCGUGGUAAUGGUGGUCGGGG---UCAU ..(((((((.(((((...))))).)))))))................((.(((((.(((((((..(...)..))))))(((....)))..).))))).---)).. ( -32.70) >DroEre_CAF1 361 105 - 1 UGGGUAGAUGGGCAGAUCCUGGUGAUCUGCCAUCGGAUUUGAGGUAGGAGUCUGAGCCUGGCUUGGAGUGUGAGUCGGCGUGGUUACGGUGGUCGGGGUGGUCAA ......(((.(.(.((((.(.((((((((((.(((.((((.(((((((........))).)))).)))).)))...)))).)))))).).))))..).).))).. ( -34.80) >consensus UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAUUGAGGUAGGAGUCUGAGCCUGGCUUGGAGUUUGAGUCGGCGUGGUAAUGGUGGUCGGGG___UCAU ..(((((((.(((((...))))).))))))).(((((((..((.((((........)))).))..)).))))).(((((............)))))......... (-30.29 = -30.60 + 0.31)

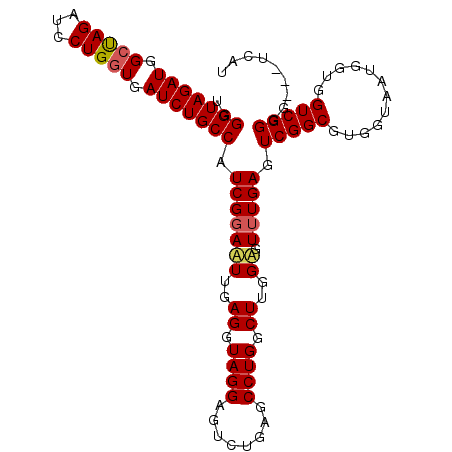
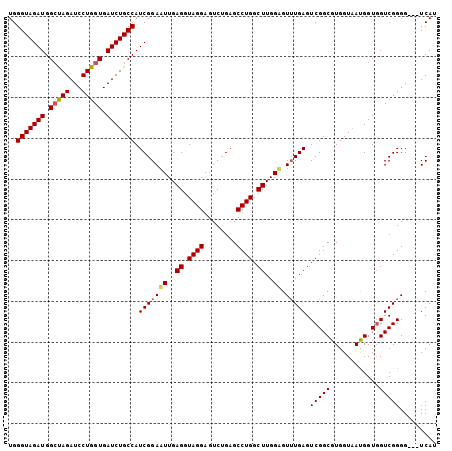
| Location | 1,510,940 – 1,511,039 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1510940 99 + 22224390 ---------CAAGCCAGGCUCAGACUCCUACCUCA------AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA---AAGCGACC---AAUGCCGCUGAAUGACCAGUAUCAAC ---------.......((.(((.............------.....(((((((((((.....))))).))))))......---.((((...---.....))))...)))))......... ( -24.30) >DroSec_CAF1 386 98 + 1 ---------CAAGCCAGGCUCCGACUCCUACCUCA------AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA----AGCGAAC---GAUGCCGCUGAACGACCAGUACCAAC ---------.......(((..((............------.....(((((((((((.....))))).))))))....(.----...)..)---)..)))((((......))))...... ( -24.20) >DroSim_CAF1 1960 99 + 1 ---------CAAGCCAGGCUCAGACUCCUACCUCA------AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA---AAGCGACC---AAUGCCGCUGAACGACCAGUACCAAC ---------.......((.((..............------.....(((((((((((.....))))).))))))......---.((((...---.....))))....))))......... ( -23.60) >DroEre_CAF1 401 102 + 1 ---------CAAGCCAGGCUCAGACUCCUACCUCA------AAUCCGAUGGCAGAUCACCAGGAUCUGCCCAUCUACCCAUCUAAGCGACC---AAUGCCGCUGAACGACCAGUAUCAAC ---------.......((.((..............------.....(((((((((((.....)))))).)))))..........((((...---.....))))....))))......... ( -23.60) >DroYak_CAF1 1820 120 + 1 CUUAUACACCAAGCCAGGCUCAGACUUCUACCUCAAACCCAAAUCCGAUGGCAGAUCACCAAGAUCUGCCCAUCUAUCCAACUAAGCGACCACUAAUGCCGCUGAACGACCAGUAUCAAC ................(((..((....)).................(((((((((((.....)))))).))))).......................)))((((......))))...... ( -23.60) >consensus _________CAAGCCAGGCUCAGACUCCUACCUCA______AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA___AAGCGACC___AAUGCCGCUGAACGACCAGUAUCAAC ................(((...((........))............(((((((((((.....))))).)))))).......................)))((((......))))...... (-20.74 = -21.14 + 0.40)

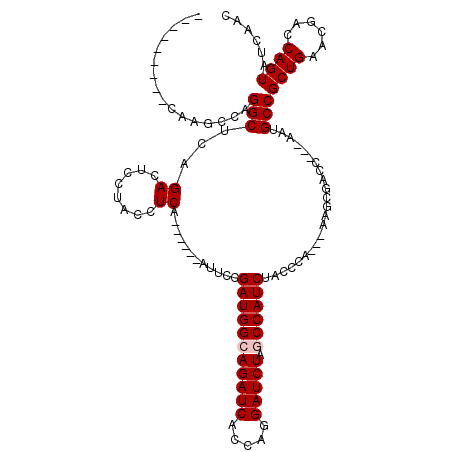
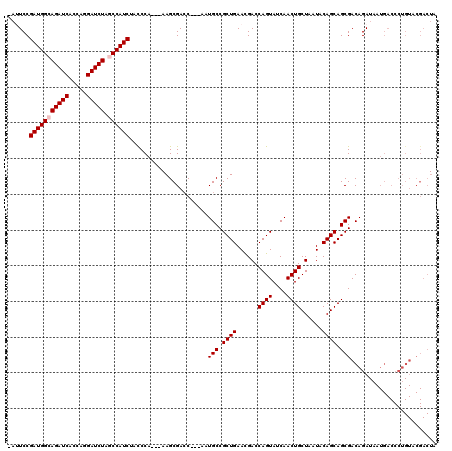
| Location | 1,510,966 – 1,511,079 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1510966 113 + 22224390 -AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA---AAGCGACC---AAUGCCGCUGAAUGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUA -.....(((((((((((.....))))).))))))......---.((((...---..(((.((((.((...((((....))))...)).)))).)))((((.........)))).)).)). ( -31.90) >DroSec_CAF1 412 112 + 1 -AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA----AGCGAAC---GAUGCCGCUGAACGACCAGUACCAACUGCUAAUACAGCAGCGACAGANNNNNNNNNNNNNNNNNNN -.....(((((((((((.....))))).))))))......----.......---(.(((.((((......((((....))))......)))).))).)...................... ( -27.40) >DroSim_CAF1 1986 113 + 1 -AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA---AAGCGACC---AAUGCCGCUGAACGACCAGUACCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUA -.....(((((((((((.....))))).))))))......---.((((...---..(((.((((......((((....))))......)))).)))((((.........)))).)).)). ( -30.80) >DroEre_CAF1 427 116 + 1 -AAUCCGAUGGCAGAUCACCAGGAUCUGCCCAUCUACCCAUCUAAGCGACC---AAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUUAUGACCCUGUACGACUA -.....(((((((((((.....)))))).)))))..........((((...---..(((.((((......((((....))))......)))).)))((((.........)))).)).)). ( -30.80) >DroYak_CAF1 1860 120 + 1 AAAUCCGAUGGCAGAUCACCAAGAUCUGCCCAUCUAUCCAACUAAGCGACCACUAAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUA ......(((((((((((.....)))))).)))))..........((((........(((.((((......((((....))))......)))).)))((((.........)))).)).)). ( -30.90) >consensus _AUUCCGAUGGCAGAUCACCAGGAUCUAGCCAUCUACCCA___AAGCGACC___AAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUA ......(((((((((((.....))))).))))))......................(((.((((......((((....))))......)))).)))........................ (-24.52 = -24.92 + 0.40)

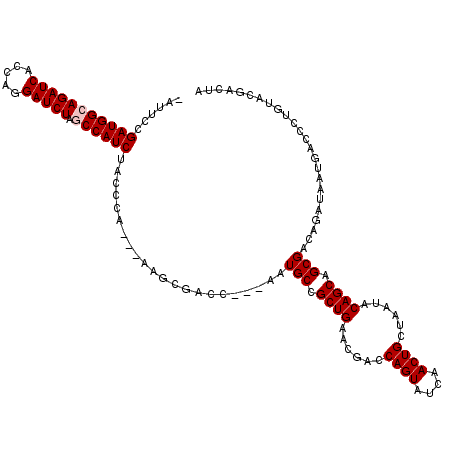
| Location | 1,510,966 – 1,511,079 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -33.64 |
| Energy contribution | -33.24 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
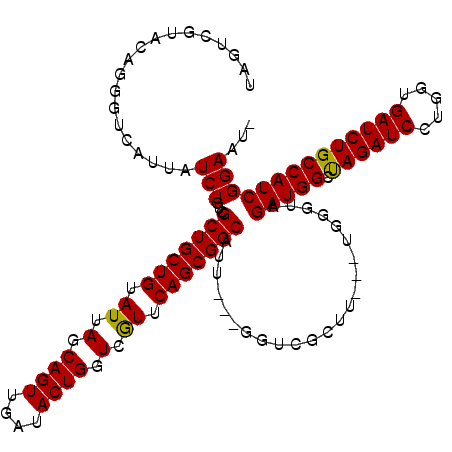
>X_DroMel_CAF1 1510966 113 - 22224390 UAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCAUUCAGCGGCAUU---GGUCGCUU---UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAU- ...(((..((((((.((((..((((((((((((....)))).((((....))))..)))))))).)---))).))))---))(((((((.(((((...))))).)))))))..)))...- ( -40.20) >DroSec_CAF1 412 112 - 1 NNNNNNNNNNNNNNNNNNNUCUGUCGCUGCUGUAUUAGCAGUUGGUACUGGUCGUUCAGCGGCAUC---GUUCGCU----UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAU- ...................((((..(((((((.((.(.((((....)))).).)).)))))))...---.......----..(((((((.(((((...))))).)))))))..))))..- ( -36.50) >DroSim_CAF1 1986 113 - 1 UAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGGUACUGGUCGUUCAGCGGCAUU---GGUCGCUU---UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAU- ...........(..((.((((....(((((((...))))))).)))).))..)((((((((((...---.)))))).---..(((((((.(((((...))))).)))))))....))))- ( -39.80) >DroEre_CAF1 427 116 - 1 UAGUCGUACAGGGUCAUAAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUU---GGUCGCUUAGAUGGGUAGAUGGGCAGAUCCUGGUGAUCUGCCAUCGGAUU- .(((((.(((((.......))))))(((((((...)))))))...((((.(((....((((((...---.))))))..))).))))(((((.((((((.....))))))))))).))))- ( -42.00) >DroYak_CAF1 1860 120 - 1 UAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUUAGUGGUCGCUUAGUUGGAUAGAUGGGCAGAUCUUGGUGAUCUGCCAUCGGAUUU ..........(((((..(((((((((((((((.((.(.((((....)))).).)).)))))))...(((....)))......))))))))((((((((.....))))))))....))))) ( -43.30) >consensus UAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUU___GGUCGCUU___UGGGUAGAUGGCUAGAUCCUGGUGAUCUGCCAUCGGAAU_ ...................(((...(((((((.((.(.((((....)))).).)).))))))).......................(((((.((((((.....))))))))))))))... (-33.64 = -33.24 + -0.40)

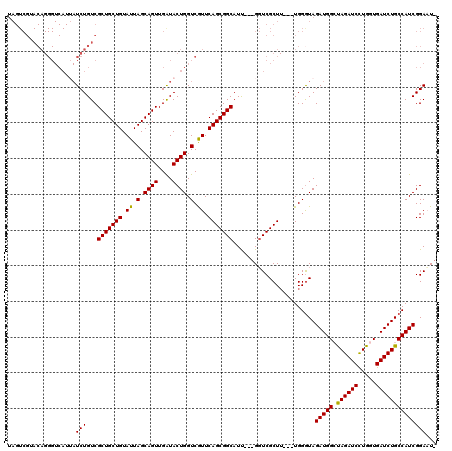
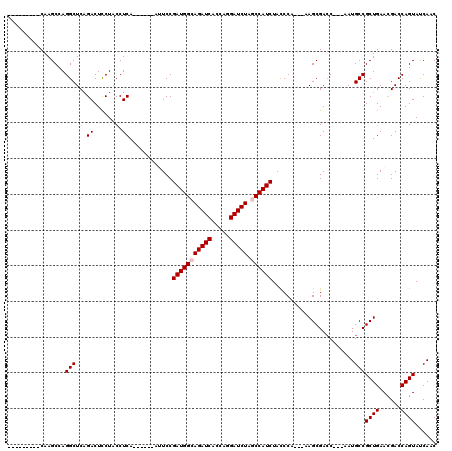
| Location | 1,511,005 – 1,511,119 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -22.60 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1511005 114 + 22224390 ---AAGCGACC---AAUGCCGCUGAAUGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAAUAGUCACAGCCACAGUCACAGU ---..(.(((.---..(((.((((.((...((((....))))...)).)))).))).............((((..(((((.(((((.....)).))).))))))))).....))).)... ( -25.30) >DroSim_CAF1 2025 110 + 1 ---AAGCGACC---AAUGCCGCUGAACGACCAGUACCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAAUAGUCACAGCCACAGUCA---- ---....(((.---..((..((((........((((...(((((.....)))))...((.....)).....))))(((((.(((((.....)).))).))))).))))..))))).---- ( -24.50) >DroEre_CAF1 466 117 + 1 UCUAAGCGACC---AAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUUAUGACCCUGUACGACUAUUACCUCUACCAGCGUAACAGUCACAGUCUCAGUCUCAGU .....(.(((.---..(((.((((......((((....))))......)))).)))........(((..((((..((((.((((((.....)).)))).))))))))..)))))).)... ( -27.20) >DroYak_CAF1 1900 120 + 1 ACUAAGCGACCACUAAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAACAGUCACAGUCAUAGUCUCAGU ................(((.((((......((((....))))......)))).)))..((((.(((((..(((..((((..(((((.....)).)))..)))))))))))).)))).... ( -24.50) >consensus ___AAGCGACC___AAUGCCGCUGAACGACCAGUAUCAACUGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAACAGUCACAGCCACAGUCACAGU .......(((......(((.((((......((((....))))......)))).))).............((((..(((((.(((((.....)).))).))))))))).....)))..... (-22.60 = -23.10 + 0.50)

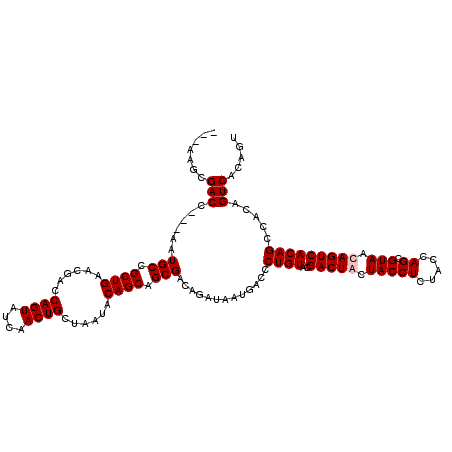
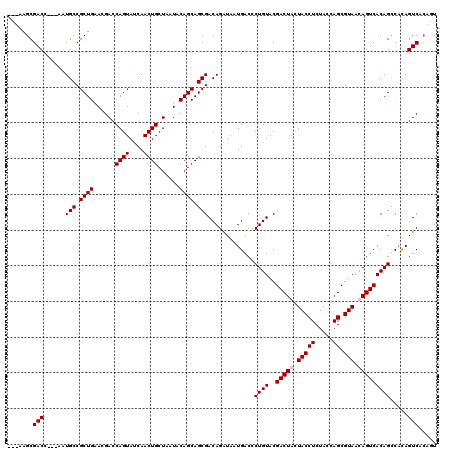
| Location | 1,511,005 – 1,511,119 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -37.38 |
| Energy contribution | -37.00 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1511005 114 - 22224390 ACUGUGACUGUGGCUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCAUUCAGCGGCAUU---GGUCGCUU--- ...((((((...((((((((((((((.((.....)))))))))))......(..((.((((....(((((((...))))))).)))).))..).....)))))...---))))))..--- ( -42.10) >DroSim_CAF1 2025 110 - 1 ----UGACUGUGGCUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGGUACUGGUCGUUCAGCGGCAUU---GGUCGCUU--- ----.....((((((..(((((((((.((.....)))))))))))(.(((((.......))))))(((((((.((.(.((((....)))).).)).)))))))...---))))))..--- ( -39.70) >DroEre_CAF1 466 117 - 1 ACUGAGACUGAGACUGUGACUGUUACGCUGGUAGAGGUAAUAGUCGUACAGGGUCAUAAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUU---GGUCGCUUAGA .(((((.....((((..(((((((((.((.....)))))))))))(.(((((.......))))))(((((((.((.(.((((....)))).).)).)))))))...---)))).))))). ( -41.50) >DroYak_CAF1 1900 120 - 1 ACUGAGACUAUGACUGUGACUGUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUUAGUGGUCGCUUAGU ((((((..((((((((..(((.((((....)))).)))..))))))))...(..(((((..((((((((....((.(.((((....)))).).)).)))))))).)))))..).)))))) ( -41.50) >consensus ACUGAGACUGUGACUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCAGUUGAUACUGGUCGUUCAGCGGCAUU___GGUCGCUU___ .........((((((..(((((((((.((.....)))))))))))(.(((((.......))))))(((((((.((.(.((((....)))).).)).)))))))......))))))..... (-37.38 = -37.00 + -0.38)

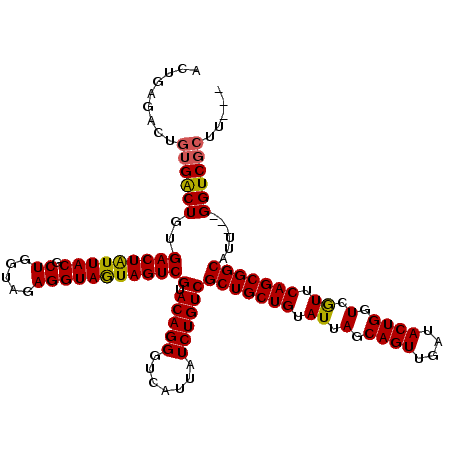
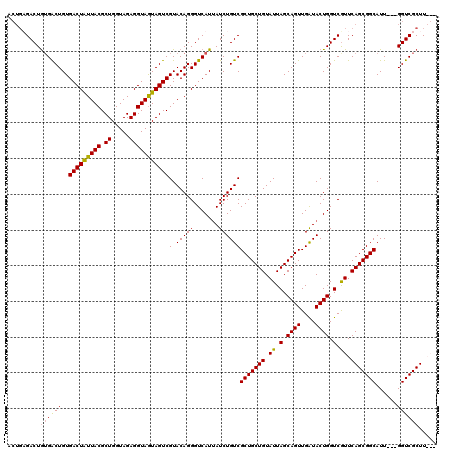
| Location | 1,511,039 – 1,511,129 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -12.50 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1511039 90 + 22224390 UGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAAUAGUCACAGCCACAGUCACAGUCACAG------UCGCA------------------------ ((((.....))))(((((.(((..((((.((((..(((((.(((((.....)).))).))))))))).....))))..)))...)------)))).------------------------ ( -25.20) >DroSim_CAF1 2059 84 + 1 UGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAAUAGUCACAGCCACAGUCA------CAG------UCGCA------------------------ ((((.....))))(((((.(((..((...((((..(((((.(((((.....)).))).))))))))).))..))).------..)------)))).------------------------ ( -21.90) >DroEre_CAF1 503 120 + 1 UGCUAAUACAGCAGCGACAGAUUAUGACCCUGUACGACUAUUACCUCUACCAGCGUAACAGUCACAGUCUCAGUCUCAGUCACAACCUCAACCACAGUCACAGCCACAACCACAACCACA ((((.....))))(.(((.((...(((..((((..((((.((((((.....)).)))).))))))))..)))...)).))).)..................................... ( -21.00) >DroYak_CAF1 1940 103 + 1 UGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAACAGUCACAGUCAUAGUCUCAGUCACAGUGUCAGUCACAAUCACGG----------------- ((((.....))))(.(((((((.(((((..(((..((((..(((((.....)).)))..)))))))))))).))))..))).).(((.....)))........----------------- ( -21.70) >consensus UGCUAAUACAGCAGCGACAGAUAAUGACCCUGUACGACUACUACCUCUACCAGCGUAACAGUCACAGCCACAGUCACAGUCACAG______UCACA________________________ ((((.....))))............(((.((((..(((((.(((((.....)).))).))))))))).....)))............................................. (-12.50 = -13.00 + 0.50)


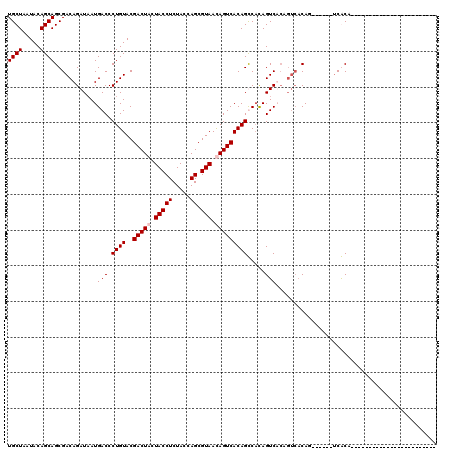
| Location | 1,511,039 – 1,511,129 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1511039 90 - 22224390 ------------------------UGCGA------CUGUGACUGUGACUGUGGCUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCA ------------------------.((((------(...((..((((((.((....((((((((((.((.....))))))))))))..)).))))))..)).))))).((((...)))). ( -34.70) >DroSim_CAF1 2059 84 - 1 ------------------------UGCGA------CUG------UGACUGUGGCUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCA ------------------------.((((------(.(------(((((.((....((((((((((.((.....))))))))))))..)).)))))).....))))).((((...)))). ( -32.00) >DroEre_CAF1 503 120 - 1 UGUGGUUGUGGUUGUGGCUGUGACUGUGGUUGAGGUUGUGACUGAGACUGAGACUGUGACUGUUACGCUGGUAGAGGUAAUAGUCGUACAGGGUCAUAAUCUGUCGCUGCUGUAUUAGCA .(..((..((((....))))..))..).((((((((.(((((.((...(((..(((((((((((((.((.....)))))))))))..))))..)))...)).))))).)))...))))). ( -40.50) >DroYak_CAF1 1940 103 - 1 -----------------CCGUGAUUGUGACUGACACUGUGACUGAGACUAUGACUGUGACUGUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCA -----------------..(((((.((((.((((.(((((.....((((((.(((...(((........)))...))).)))))).))))).))))))))..))))).((((...)))). ( -30.80) >consensus ________________________UGCGA______CUGUGACUGAGACUGUGACUGUGACUAUUACGCUGGUAGAGGUAGUAGUCGUACAGGGUCAUUAUCUGUCGCUGCUGUAUUAGCA .....................................(((((...((..((((((.((((((((((.((.....)))))))))))....).))))))..)).))))).((((...)))). (-22.48 = -22.85 + 0.37)

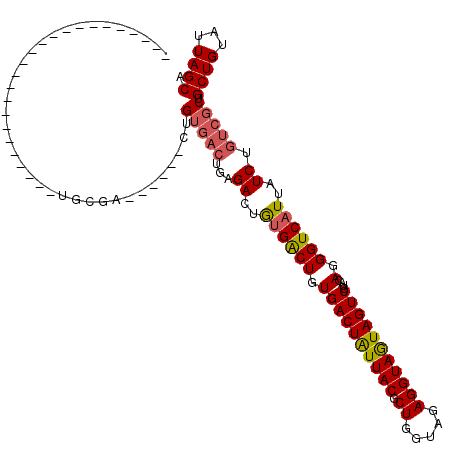
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:43 2006