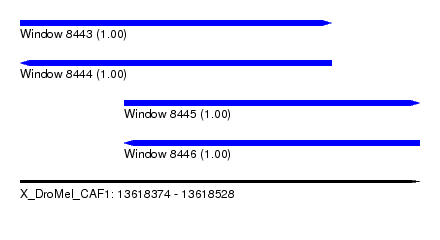
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,618,374 – 13,618,528 |
| Length | 154 |
| Max. P | 0.999824 |

| Location | 13,618,374 – 13,618,494 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -54.40 |
| Consensus MFE | -52.42 |
| Energy contribution | -51.50 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13618374 120 + 22224390 UACCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAAGGCUGCGCCGCCCACGCAGGGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGC ((((.((........((((((((((....)).))))))))((((((((((((.....)))....)))))))))(((((......))))))).))))....((((.(......).)))).. ( -54.40) >DroSec_CAF1 4246 120 + 1 UACCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAAGGCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGC .....((........((((((((((....)).))))))))((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))))) ( -56.20) >DroSim_CAF1 4222 120 + 1 UACCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAAGGCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGC .....((........((((((((((....)).))))))))((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))))) ( -56.20) >DroEre_CAF1 4197 118 + 1 --CCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAGGGUUGCGCCGCCCACGUAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGUCGCCAAGGCCGUCGGCGC --...((........((((((((((....)).))))))))((((((.......))))))(((((((((...(((((...((((((((...))))))))..)))))....))))))))))) ( -51.70) >DroYak_CAF1 3983 118 + 1 --CCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAAGGCUGUGCCGCCAACGCAGCGUCGGUGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGC --...((........((((((((((....)).))))))))(((((((((((.(((...))).)))))))))))(((((......)))))(((((......(((.....)))..))))))) ( -53.50) >consensus UACCAGCAUCAUCGACUUAAUCCGUUCGAGCGGGAUUAAGGCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGC .....((........((((((((((....)).))))))))((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))))) (-52.42 = -51.50 + -0.92)


| Location | 13,618,374 – 13,618,494 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -57.60 |
| Consensus MFE | -53.86 |
| Energy contribution | -54.90 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13618374 120 - 22224390 GCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACCCUGCGUGGGCGGCGCAGCCUUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGGUA .((((((.....))))))......((((((((((((......)))))(((((((((((.((.....)).)))))))))))((((((((.........))))))))........))))))) ( -59.80) >DroSec_CAF1 4246 120 - 1 GCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGCCUUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGGUA .((((((.....))))))......((((((((((((......)))))(((((((((((.(((...))).)))))))))))((((((((.........))))))))........))))))) ( -61.20) >DroSim_CAF1 4222 120 - 1 GCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGCCUUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGGUA .((((((.....))))))......((((((((((((......)))))(((((((((((.(((...))).)))))))))))((((((((.........))))))))........))))))) ( -61.20) >DroEre_CAF1 4197 118 - 1 GCGCCGACGGCCUUGGCGACUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUACGUGGGCGGCGCAACCCUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGG-- (((.((.((((...(((((((.((((((((...))))))))..)))))))((((((((.(((...))).)))))))).....((((((.........)))))).)))).)).)))...-- ( -53.70) >DroYak_CAF1 3983 118 - 1 GCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCACCGACGCUGCGUUGGCGGCACAGCCUUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGG-- .((((((.....))))))........((((((((((......)))))((((.(((.((((((...)))))).))).))))((((((((.........))))))))........)))))-- ( -52.10) >consensus GCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGCCUUAAUCCCGCUCGAACGGAUUAAGUCGAUGAUGCUGGUA .((((((.....)))))).......(((((((((((......)))))(((((((((((.((.....)).)))))))))))((((((((.........))))))))........)))))). (-53.86 = -54.90 + 1.04)


| Location | 13,618,414 – 13,618,528 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -50.33 |
| Consensus MFE | -41.82 |
| Energy contribution | -41.06 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13618414 114 + 22224390 GCUGCGCCGCCCACGCAGGGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGCUACACAGAAAGAAAUGUAAAGG------UAGAAUUGGUCU ((((((((((((.....)))....)))))))))(((((..(((((((.((((((......(((.....)))..))))))))..(((........)))...))------)))...))))). ( -48.70) >DroSec_CAF1 4286 114 + 1 GCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGCUACACAGAAAGAAAUGUACAGG------UAGAAGUGACCU ((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))).((((......(....)......)------)))......... ( -48.90) >DroSim_CAF1 4262 114 + 1 GCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGCUACACAGAAAGAAAUGUACAGG------UAGAAGUGACCU ((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))).((((......(....)......)------)))......... ( -48.90) >DroEre_CAF1 4235 114 + 1 GUUGCGCCGCCCACGUAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGUCGCCAAGGCCGUCGGCGCUACACAGAAAGAAAAGUAAAAG------UAGAAGGGGCCU ........((((..((((((((((((((...(((((...((((((((...))))))))..)))))....))))))))))))))((..........)).....------......)))).. ( -52.90) >DroYak_CAF1 4021 120 + 1 GCUGUGCCGCCAACGCAGCGUCGGUGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGCUACACUGCAAGAAAAGUAAAAGUGAAAGUAAAGGUGGCCU (((((((((((.(((...))).)))))))))))(((((......)))))(.((((.....)))).).(((((..(...(((.((((..............))))..)))...)..))))) ( -52.24) >consensus GCUGCGCCGCCCACGCAGCGUCGACGGCGCAGCGACCGCAUUACCGGUCGCCGGUAAUUAGCCGCCAAGGCCGUCGGCGCUACACAGAAAGAAAUGUAAAGG______UAGAAGUGGCCU ((((((.......))))))(((((((((...(((((((......)))))))((((.....)))).....))))))))).......................................... (-41.82 = -41.06 + -0.76)



| Location | 13,618,414 – 13,618,528 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -52.66 |
| Consensus MFE | -47.46 |
| Energy contribution | -47.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13618414 114 - 22224390 AGACCAAUUCUA------CCUUUACAUUUCUUUCUGUGUAGCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACCCUGCGUGGGCGGCGCAGC .((((.......------...((((((........))))))((((((.....))))))((..((((((((...))))))))))))))(((((((((((.((.....)).))))))))))) ( -53.60) >DroSec_CAF1 4286 114 - 1 AGGUCACUUCUA------CCUGUACAUUUCUUUCUGUGUAGCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGC ((((.......)------)))........((.((..(((((((.(((((((...(((((((.((((((((...))))))))..)))))))..))))))).)))))))..)).))...... ( -53.60) >DroSim_CAF1 4262 114 - 1 AGGUCACUUCUA------CCUGUACAUUUCUUUCUGUGUAGCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGC ((((.......)------)))........((.((..(((((((.(((((((...(((((((.((((((((...))))))))..)))))))..))))))).)))))))..)).))...... ( -53.60) >DroEre_CAF1 4235 114 - 1 AGGCCCCUUCUA------CUUUUACUUUUCUUUCUGUGUAGCGCCGACGGCCUUGGCGACUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUACGUGGGCGGCGCAAC ..((.((.((((------(....((..........))((((((.(((((((...(((((((.((((((((...))))))))..)))))))..))))))).))))))))))).)).))... ( -55.70) >DroYak_CAF1 4021 120 - 1 AGGCCACCUUUACUUUCACUUUUACUUUUCUUGCAGUGUAGCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCACCGACGCUGCGUUGGCGGCACAGC ..(((.((...((..........(((........)))((((((.((..(((...(((((((.((((((((...))))))))..)))))))..)))..)).)))))))).)).)))..... ( -46.80) >consensus AGGCCACUUCUA______CCUUUACAUUUCUUUCUGUGUAGCGCCGACGGCCUUGGCGGCUAAUUACCGGCGACCGGUAAUGCGGUCGCUGCGCCGUCGACGCUGCGUGGGCGGCGCAGC .............................((.((..(((((((.(((((((...(((((((.((((((((...))))))))..)))))))..))))))).)))))))..)).))...... (-47.46 = -47.94 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:22 2006