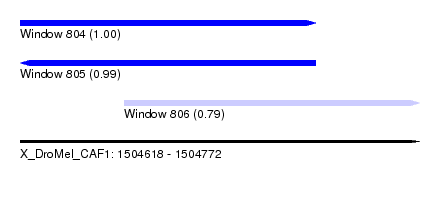
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,504,618 – 1,504,772 |
| Length | 154 |
| Max. P | 0.998856 |

| Location | 1,504,618 – 1,504,732 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -52.77 |
| Consensus MFE | -31.54 |
| Energy contribution | -34.48 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1504618 114 + 22224390 CAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGUGGGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGC ..((((...((((.(((.....((.((((......)))).))...(((((((((..(((...)))..))).((((((((....))))))))..)))))).))).))))..)))) ( -57.30) >DroVir_CAF1 48029 96 + 1 CUGCGGCGG---CGGUUAACAAUAGUGUU------GCAGGG---------GGCGGGGCUGGGGGGAAUGCUGUGGCCGCAGCAGCAGCUGCCGCUGCGGCCGCUGCUGAGCUGC (((((((.(---((((...(..((((.((------((....---------.)))).))))..).....))))).)))))))(((((((.((((...)))).)))))))...... ( -51.20) >DroSec_CAF1 29326 114 + 1 CAGCAGCAGCGGCGGCCAACAAUAGCGUUAAUGUAGGCGGUGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGC ..((((...((((.(((.....((.((((......)))).))...(((((((((..(((...)))..))).((((((((....))))))))..)))))).))).))))..)))) ( -57.20) >DroWil_CAF1 22074 90 + 1 CUGCUGCUG---------GUAAUAGUG------UUGGCGGCG---------GUGGGGCCAAUGGCAAUGCUGUGGCAGCGGCAGCUGCAGCAGCUGCUGCCGCCGCUGAGUUAU (..((((((---------.((((...)------))).)))))---------)..).(((...)))(((...(((((.((((((((((...)))))))))).)))))...))).. ( -45.40) >consensus CAGCAGCAG___CGGCCAACAAUAGCGUU____U_GGCGGUG________GGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCAGCAGCCGCAGCUGCCGCCGCGGCCGAACUGC .....((((...(((((..................................(((..(((...)))..))).((((((((((((((....)))))))))))))))))))..)))) (-31.54 = -34.48 + 2.94)

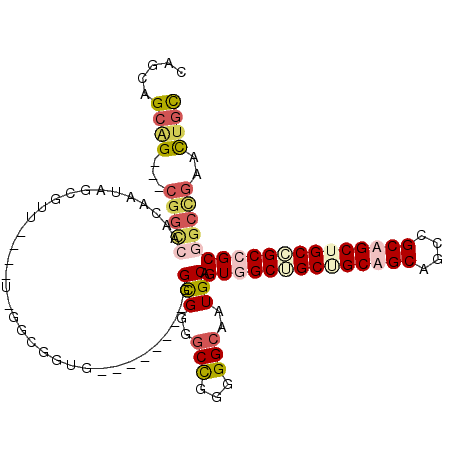
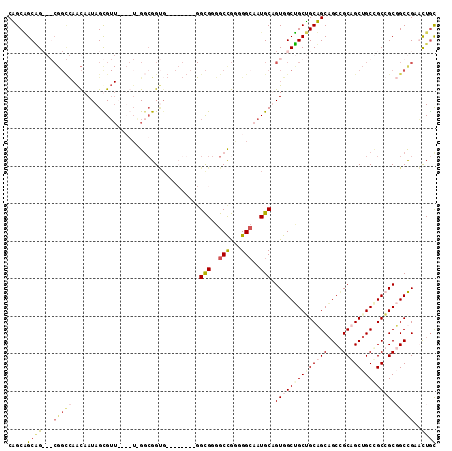
| Location | 1,504,618 – 1,504,732 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -26.38 |
| Energy contribution | -27.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1504618 114 - 22224390 GCAGUUCGGCCGCCGCGGCAGCUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCCCACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUG (((((.((((.((((.(((((.(((((((((....))))))...))))))))..))))..................(((.(((.((....)).))).))).))))..))))).. ( -48.00) >DroVir_CAF1 48029 96 - 1 GCAGCUCAGCAGCGGCCGCAGCGGCAGCUGCUGCUGCGGCCACAGCAUUCCCCCCAGCCCCGCC---------CCCUGC------AACACUAUUGUUAACCG---CCGCCGCAG ((.((...((((.((((((((((((....))))))))))))...((..........))......---------..))))------((((....)))).....---..)).)).. ( -38.80) >DroSec_CAF1 29326 114 - 1 GCAGUUCGGCCGCCGCGGCAGCUGCGGCUGCAGCAGCAGCCACUGCAUUGCCCCCGGCCCCGCCUACCGAUCCACCGCCUACAUUAACGCUAUUGUUGGCCGCCGCUGCUGCUG (((((.((((.((((.(((((.(((((((((....))))))...))))))))..))))..................(((.(((.((....)).))).))).))))..))))).. ( -48.00) >DroWil_CAF1 22074 90 - 1 AUAACUCAGCGGCGGCAGCAGCUGCUGCAGCUGCCGCUGCCACAGCAUUGCCAUUGGCCCCAC---------CGCCGCCAA------CACUAUUAC---------CAGCAGCAG ........((((((((((..((((..(((((....)))))..)))).)))))...((.....)---------))))))...------.........---------......... ( -33.10) >consensus GCAGCUCAGCCGCCGCGGCAGCUGCGGCUGCAGCAGCAGCCACAGCAUUGCCCCCGGCCCCGCC________CACCGCC_A____AACACUAUUGUUGGCCG___CUGCUGCAG ......(((((((.((....)).)))))))((((((((((....((...((.....))...)).....................(((((....)))))......)))))))))) (-26.38 = -27.50 + 1.12)

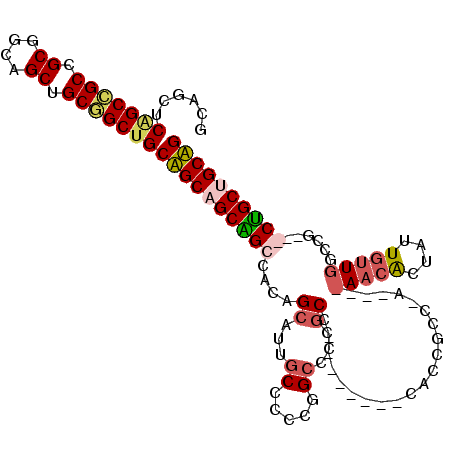
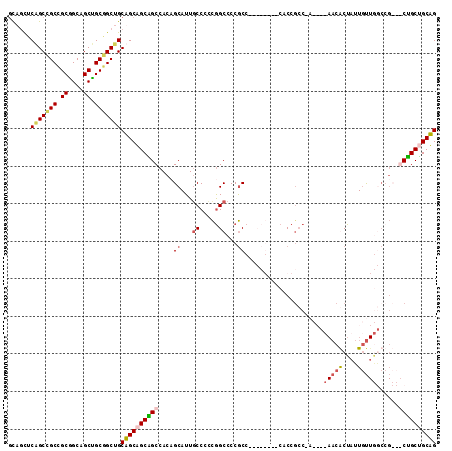
| Location | 1,504,658 – 1,504,772 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -60.63 |
| Consensus MFE | -42.51 |
| Energy contribution | -44.97 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1504658 114 + 22224390 UGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCCGUGGCCGGGGG .((..((((((.(((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))....)))))(....)..))).)))))))))...))..... ( -63.60) >DroSec_CAF1 29366 114 + 1 UGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCCGUGGCCGGGGG .((..((((((.(((.((((((((....((((((((((((((.(((((....))))).)))))))))..)))))....)))))(....)..))).)))))))))...))..... ( -63.60) >DroWil_CAF1 22099 105 + 1 CG---------GUGGGGCCAAUGGCAAUGCUGUGGCAGCGGCAGCUGCAGCAGCUGCUGCCGCCGCUGAGUUAUUGCUCUCACCAAUUGUGGGGGCAGCCGCUGUGGCGGGGGG ((---------(((((((....((((..(((((.(((((....))))).)))))))))))).)))))).....((((((((((.....))))))))))((((....)))).... ( -54.70) >consensus UGGAUCGGUAGGCGGGGCCGGGGGCAAUGCAGUGGCUGCUGCUGCAGCCGCAGCUGCCGCGGCGGCCGAACUGCUUCUCUCUCCGAUCGUUGGCGCCGCUGCCGUGGCCGGGGG ....(((((.((((((((((((((....((((((((((((((.(((((....))))).)))))))))..))))).....)))))).........))).)))))...)))))... (-42.51 = -44.97 + 2.46)

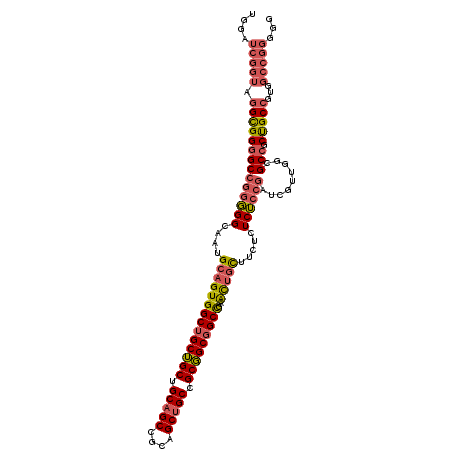
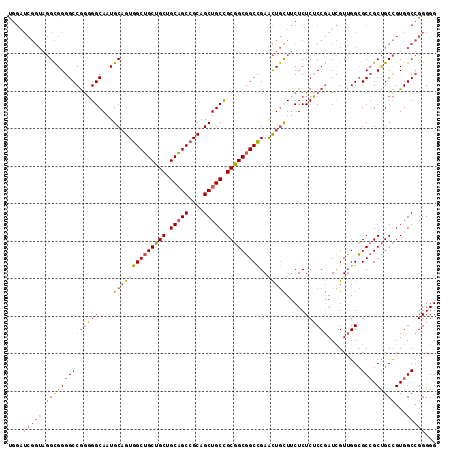
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:36 2006