
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,587,720 – 13,587,817 |
| Length | 97 |
| Max. P | 0.969122 |

| Location | 13,587,720 – 13,587,817 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
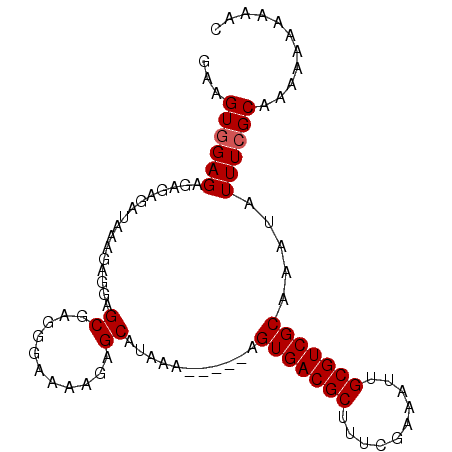
>X_DroMel_CAF1 13587720 97 + 22224390 GAAGUGGAGAGAGAGAUAAAGAGGAGCGAUGGAAAAGAGCAUAAAAAUAAAGUGACGCUGUCGAAAUUGCGUCGCAAACAUUUCGCAAAAAAUAAAA ((((((...................((..(.....)..))...........(((((((..........)))))))...))))))............. ( -19.30) >DroSec_CAF1 3459 91 + 1 GAAGUGGAGAGAGAGAUAAAGAGGAGCGAGGGAAAAGAGCAUAAA-----AGUGACGCUUUCGAAAUUGCGUCGCAAAUAUUUCGCAAAAA-AAAAC ...((((((................((...........)).....-----.(((((((..........))))))).....)))))).....-..... ( -17.80) >DroSim_CAF1 3632 92 + 1 GAAGUUGAGAGAGAGAUAAAGAGGAGCGAGGGAAAAGAGCAUAAA-----AGUGACGCUUUCGAAAUUGCGUCGCAAAUAUUUCGCAAAAAAAAAAC ..........(.((((((.......((...........)).....-----.(((((((..........)))))))...)))))).)........... ( -17.30) >consensus GAAGUGGAGAGAGAGAUAAAGAGGAGCGAGGGAAAAGAGCAUAAA_____AGUGACGCUUUCGAAAUUGCGUCGCAAAUAUUUCGCAAAAAAAAAAC ...((((((................((...........))...........(((((((..........))))))).....))))))........... (-16.50 = -16.83 + 0.33)

| Location | 13,587,720 – 13,587,817 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -10.57 |
| Consensus MFE | -9.90 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
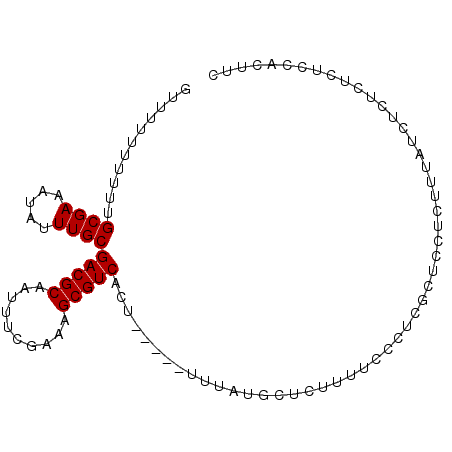
>X_DroMel_CAF1 13587720 97 - 22224390 UUUUAUUUUUUGCGAAAUGUUUGCGACGCAAUUUCGACAGCGUCACUUUAUUUUUAUGCUCUUUUCCAUCGCUCCUCUUUAUCUCUCUCUCCACUUC ...........((((...(....)(((((..........)))))........................))))......................... ( -11.10) >DroSec_CAF1 3459 91 - 1 GUUUU-UUUUUGCGAAAUAUUUGCGACGCAAUUUCGAAAGCGUCACU-----UUUAUGCUCUUUUCCCUCGCUCCUCUUUAUCUCUCUCUCCACUUC .....-.....((((.(((...(.(((((..........))))).).-----..)))...........))))......................... ( -10.30) >DroSim_CAF1 3632 92 - 1 GUUUUUUUUUUGCGAAAUAUUUGCGACGCAAUUUCGAAAGCGUCACU-----UUUAUGCUCUUUUCCCUCGCUCCUCUUUAUCUCUCUCUCAACUUC ...........((((.(((...(.(((((..........))))).).-----..)))...........))))......................... ( -10.30) >consensus GUUUUUUUUUUGCGAAAUAUUUGCGACGCAAUUUCGAAAGCGUCACU_____UUUAUGCUCUUUUCCCUCGCUCCUCUUUAUCUCUCUCUCCACUUC ...........((((.....))))(((((..........)))))..................................................... ( -9.90 = -9.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:08 2006