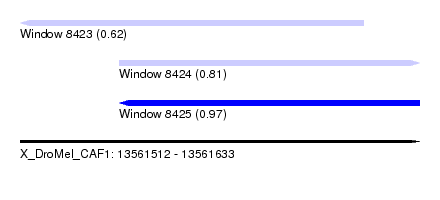
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,561,512 – 13,561,633 |
| Length | 121 |
| Max. P | 0.968933 |

| Location | 13,561,512 – 13,561,616 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.59 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -15.02 |
| Energy contribution | -16.52 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13561512 104 - 22224390 CCAAUUGCGGCUUAAAACAUGAAG--AAAGCUGCAAAAAAUCGAAAAUGAAUAUGAGAAACAGAUCAACUGGCAACCAGAAUGCCAUGCGAUGCAGAUCGAUUCCU ....(((((((((...........--.)))))))))....................(((...((((...(((((.......)))))(((...)))))))..))).. ( -22.20) >DroSec_CAF1 16435 106 - 1 CCAAUUGCGGCUUAAAACAUGAAGAGAAAGCUGCAAAGAAUCGGAAAUGAAUAUGAGAAAGAGAUCAACUGGCAAUCAGAAUGCCAUGCGAUGCAGAUCGAUUCCU ....(((((((((..............))))))))).(((((((...((....(((........)))..(((((.......))))).......))..))))))).. ( -24.54) >DroSim_CAF1 8868 106 - 1 CCAAUUGCGGCUUAAAACAUGAAGAGAAAGCUGCAAAGAAUCGGAAAUGAAUUUGAGAAACAGAUCAACUGGCAAUCAGAAUGCCAUGCGAUGCAGAUCGAUUCCU ....(((((((((..............))))))))).(((((((...(((.((((.....)))))))..(((((.......)))))...........))))))).. ( -25.64) >DroYak_CAF1 16246 101 - 1 CCAAUUGCGGCUUAAAACAUGAAG--AAAGCUACAAAAGG---UAAAGAAAUAUGCAAAAAUUAUCAACUGCAAAUCAGAAUCCAAUGCAAUGCAGUUCGAUUUCU ((..(((..((((...........--.))))..)))..))---....................((((((((((...((........))...))))))).))).... ( -16.00) >consensus CCAAUUGCGGCUUAAAACAUGAAG__AAAGCUGCAAAAAAUCGGAAAUGAAUAUGAGAAACAGAUCAACUGGCAAUCAGAAUGCCAUGCGAUGCAGAUCGAUUCCU ....(((((((((..............))))))))).(((((((.........................(((((.......)))))(((...)))..))))))).. (-15.02 = -16.52 + 1.50)



| Location | 13,561,542 – 13,561,633 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13561542 91 + 22224390 GUUGCCAGUUGAUCUGUUUCUCAUAUUCAUUUUCGAUUUUUUGCAGCUUU--CUUCAUGUUUUAAGCCGCAAUUGGCAGACUGGCGGCAUUUU ((((((((....((((((........((......))....((((.((((.--...........)))).))))..))))))))))))))..... ( -23.30) >DroSec_CAF1 16465 93 + 1 AUUGCCAGUUGAUCUCUUUCUCAUAUUCAUUUCCGAUUCUUUGCAGCUUUCUCUUCAUGUUUUAAGCCGCAAUUGGCAGACUGGCGGCAUUUU .(((((((((((......))............(((((....(((.((((...(.....)....)))).))))))))..)))))))))...... ( -20.70) >DroSim_CAF1 8898 93 + 1 AUUGCCAGUUGAUCUGUUUCUCAAAUUCAUUUCCGAUUCUUUGCAGCUUUCUCUUCAUGUUUUAAGCCGCAAUUGGCAGACUGGCGGCAUUUU .(((((((((((......))............(((((....(((.((((...(.....)....)))).))))))))..)))))))))...... ( -20.70) >DroYak_CAF1 16276 88 + 1 AUUUGCAGUUGAUAAUUUUUGCAUAUUUCUUUA---CCUUUUGUAGCUUU--CUUCAUGUUUUAAGCCGCAAUUGGCAGACUGGCGGCAUUUU ...(((((..........)))))..........---..............--.............(((((....(.....)..)))))..... ( -13.10) >consensus AUUGCCAGUUGAUCUGUUUCUCAUAUUCAUUUCCGAUUCUUUGCAGCUUU__CUUCAUGUUUUAAGCCGCAAUUGGCAGACUGGCGGCAUUUU .(((((((((((......))................((..((((.((((...(.....)....)))).))))..))..)))))))))...... (-16.17 = -15.92 + -0.25)



| Location | 13,561,542 – 13,561,633 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
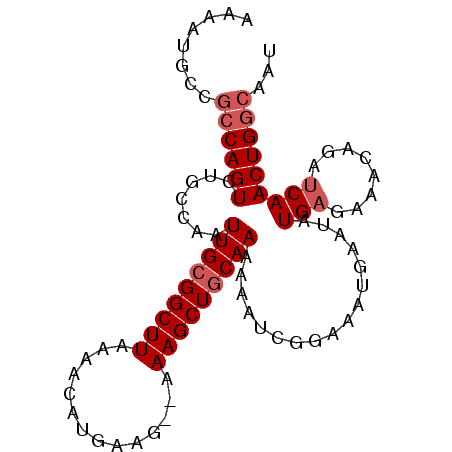
>X_DroMel_CAF1 13561542 91 - 22224390 AAAAUGCCGCCAGUCUGCCAAUUGCGGCUUAAAACAUGAAG--AAAGCUGCAAAAAAUCGAAAAUGAAUAUGAGAAACAGAUCAACUGGCAAC ........((((((.......(((((((((...........--.))))))))).................(((........)))))))))... ( -21.70) >DroSec_CAF1 16465 93 - 1 AAAAUGCCGCCAGUCUGCCAAUUGCGGCUUAAAACAUGAAGAGAAAGCUGCAAAGAAUCGGAAAUGAAUAUGAGAAAGAGAUCAACUGGCAAU ........(((((((((....(((((((((..............))))))))).....))))........(((........))).)))))... ( -22.74) >DroSim_CAF1 8898 93 - 1 AAAAUGCCGCCAGUCUGCCAAUUGCGGCUUAAAACAUGAAGAGAAAGCUGCAAAGAAUCGGAAAUGAAUUUGAGAAACAGAUCAACUGGCAAU ........(((((((((....(((((((((..............))))))))).....))))..(((.((((.....))))))).)))))... ( -24.04) >DroYak_CAF1 16276 88 - 1 AAAAUGCCGCCAGUCUGCCAAUUGCGGCUUAAAACAUGAAG--AAAGCUACAAAAGG---UAAAGAAAUAUGCAAAAAUUAUCAACUGCAAAU ....(((......((((((..(((..((((...........--.))))..)))..))---)..))).....)))................... ( -12.70) >consensus AAAAUGCCGCCAGUCUGCCAAUUGCGGCUUAAAACAUGAAG__AAAGCUGCAAAAAAUCGGAAAUGAAUAUGAGAAACAGAUCAACUGGCAAU ........((((((.......(((((((((..............))))))))).................(((........)))))))))... (-16.14 = -17.14 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:03 2006