
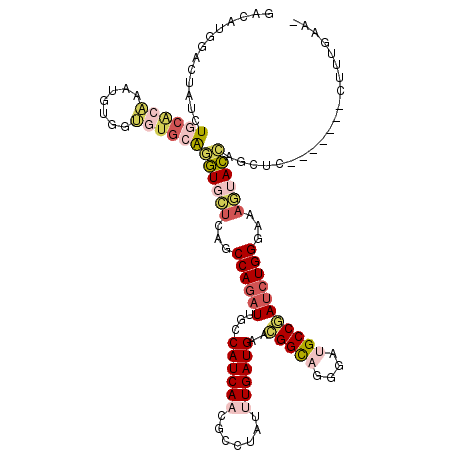
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,560,457 – 13,560,569 |
| Length | 112 |
| Max. P | 0.821520 |

| Location | 13,560,457 – 13,560,569 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -22.93 |
| Energy contribution | -24.47 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
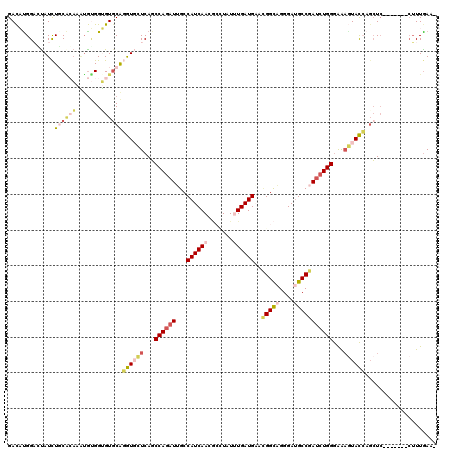
>X_DroMel_CAF1 13560457 112 + 22224390 GACAUGGAGUAUCUUCACAAAUGUGGUGUGCAGGUGCUCAGCCAGAUUGCCAUCAACGCAUAUUUGAUGAACGGCAGGGAUGCCGAUCUGGGAAAGUACCAGCUC-------CUUUGAA- ..((.(((((....((((....))))......((((((...((((((...((((((.......))))))..(((((....)))))))))))...)))))).))))-------)..))..- ( -39.50) >DroSec_CAF1 15406 112 + 1 GACAUGGAGUAUCUCCACAAAUGUGGUGUGCAGGUGCUCAGCCAGAUUGCCAUCAACGCGUAUUUGAUGAACGGCAGGGAUGCCGAUCUGGGAAAGUACCAGCUG-------CUUUGAA- .....((((((.(.((((....)))).).((.((((((...((((((...((((((.......))))))..(((((....)))))))))))...)))))).))))-------))))...- ( -39.10) >DroEre_CAF1 15384 112 + 1 GACAUGGACUAUCUGCACAAAUGUGGUGUCCAGGUGCUCAGCCAGAUUGCCAUCAACGCCUAUUUGAUGAACGGCAGGGAUGCCGAUCUGGGAAAGUACCACCUC-------CUUUAAA- ....(((((...(..((....))..).)))))((((((...((((((((.((((...(((............)))...)))).))))))))...)))))).....-------.......- ( -40.00) >DroYak_CAF1 15279 112 + 1 GACACGGAAUAUCUGCGCAAAUGUGGUGUGCAGGUGCUCAGCCAGAUUGCCAUCAACGCCUAUUUGAUGCACGGCAGGGAUGCCGAUCUGGGAAAGUACCAGCUC-------CUUUGAA- .....(((.....((((((.......))))))((((((...(((((((((.(((((.......))))))))(((((....)))))))))))...))))))...))-------)......- ( -39.10) >DroAna_CAF1 15714 112 + 1 GAUCUGGCCUAUCUGCUGAAGAAGGGCGUGCGAGUCCUCAGCCAGAUAGCCAUCAACAGUUAUCUGAUGAAGGGCCGGGAUGCCGAUCUGGGGGCCUACGAGCUC-------AGUUGAA- ..((((((((.((.(((((....((((......)))))))))((((((((........))))))))..)).))))))))....(((.(((((..(....)..)))-------)))))..- ( -40.50) >DroPer_CAF1 15396 120 + 1 GACUUGGCCCAUUUGCAGCACUGCGGGGUGCGCAUCUUCACCCAAUUGGCCAUCAACUACUAUCUGAUGCAUGGUCGGGAAGCCAAUCUGGAGAAAUAUGUGGUCAAGUAGACUUUGAAC .(((((((((((.(((.(((((....))))))))((((((.......((((((((.(........).)).))))))((....))....))))))...))).))))))))........... ( -38.30) >consensus GACAUGGACUAUCUGCACAAAUGUGGUGUGCAGGUGCUCAGCCAGAUUGCCAUCAACGCCUAUUUGAUGAACGGCAGGGAUGCCGAUCUGGGAAAGUACCAGCUC_______CUUUGAA_ .............((((((.......))))))((((((...((((((...((((((.......))))))..(((((....)))))))))))...)))))).................... (-22.93 = -24.47 + 1.53)

| Location | 13,560,457 – 13,560,569 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -22.19 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
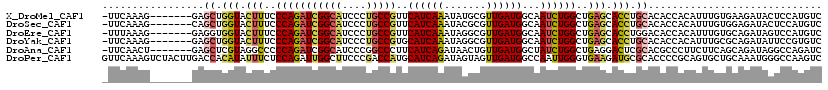
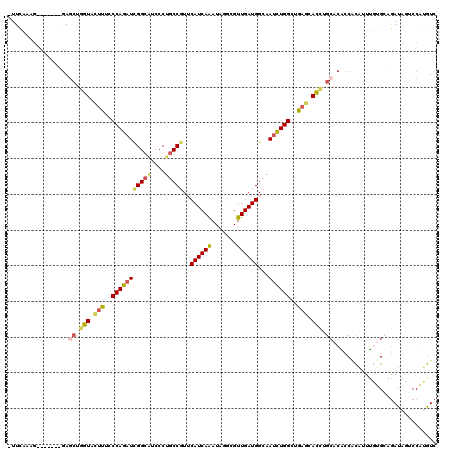
>X_DroMel_CAF1 13560457 112 - 22224390 -UUCAAAG-------GAGCUGGUACUUUCCCAGAUCGGCAUCCCUGCCGUUCAUCAAAUAUGCGUUGAUGGCAAUCUGGCUGAGCACCUGCACACCACAUUUGUGAAGAUACUCCAUGUC -......(-------((((.(((.(((..(((((((((((....))))).(((((((.......)))))))..))))))..))).))).))....(((....))).......)))..... ( -34.60) >DroSec_CAF1 15406 112 - 1 -UUCAAAG-------CAGCUGGUACUUUCCCAGAUCGGCAUCCCUGCCGUUCAUCAAAUACGCGUUGAUGGCAAUCUGGCUGAGCACCUGCACACCACAUUUGUGGAGAUACUCCAUGUC -.......-------..((.(((.(((..(((((((((((....))))).(((((((.......)))))))..))))))..))).))).))...........((((((...))))))... ( -35.30) >DroEre_CAF1 15384 112 - 1 -UUUAAAG-------GAGGUGGUACUUUCCCAGAUCGGCAUCCCUGCCGUUCAUCAAAUAGGCGUUGAUGGCAAUCUGGCUGAGCACCUGGACACCACAUUUGUGCAGAUAGUCCAUGUC -.......-------.(((((...(((..((((((.(.((((..((((............))))..)))).).))))))..))))))))((((..(((....)))......))))..... ( -34.70) >DroYak_CAF1 15279 112 - 1 -UUCAAAG-------GAGCUGGUACUUUCCCAGAUCGGCAUCCCUGCCGUGCAUCAAAUAGGCGUUGAUGGCAAUCUGGCUGAGCACCUGCACACCACAUUUGCGCAGAUAUUCCGUGUC -......(-------(((..(((.(((..((((((.(.((((..((((............))))..)))).).))))))..))).)))(((.((.......)).)))....))))..... ( -34.00) >DroAna_CAF1 15714 112 - 1 -UUCAACU-------GAGCUCGUAGGCCCCCAGAUCGGCAUCCCGGCCCUUCAUCAGAUAACUGUUGAUGGCUAUCUGGCUGAGGACUCGCACGCCCUUCUUCAGCAGAUAGGCCAGAUC -.......-------.........((((..((((((((....)))(((..(((.(((....))).))).))).)))))((((((((...(....)...)))))))).....))))..... ( -34.90) >DroPer_CAF1 15396 120 - 1 GUUCAAAGUCUACUUGACCACAUAUUUCUCCAGAUUGGCUUCCCGACCAUGCAUCAGAUAGUAGUUGAUGGCCAAUUGGGUGAAGAUGCGCACCCCGCAGUGCUGCAAAUGGGCCAAGUC ...........(((((.((.(((.((((.((.((((((((...((((..(((........)))))))..)))))))).)).)))).(((((((......)))).))).))))).))))). ( -36.50) >consensus _UUCAAAG_______GAGCUGGUACUUUCCCAGAUCGGCAUCCCUGCCGUUCAUCAAAUAGGCGUUGAUGGCAAUCUGGCUGAGCACCUGCACACCACAUUUGUGCAGAUAGUCCAUGUC .................((.(((.(((..(((((((((((....)))))..((((((.......))))))...))))))..))).))).))............................. (-22.19 = -22.20 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:00 2006