
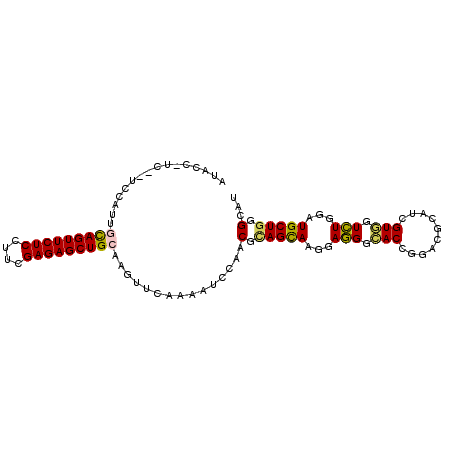
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,547,121 – 13,547,217 |
| Length | 96 |
| Max. P | 0.676632 |

| Location | 13,547,121 – 13,547,217 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13547121 96 + 22224390 ---CC-CCAACCCAUUGCAGUUCUCCUUCGAGAGCUGCAAGUUCAAAAUUCAGCGCAGCAAGGAGGGCACCGGACGUAUCGUGGUCUGGAUGCUGGGCAU ---.(-((...((.(((((((((((....)))))))))))............((...))..))..((((((((((........)))))).)))))))... ( -36.50) >DroVir_CAF1 1320 95 + 1 AUAUU-UC--UCCAU--CAGUUCUCGUUUGAGAGCUGUAAGUUUAAGAUCCAACGUAGCAAGGAAGGGACAGGACGCAUUGUUGUUUGGAUGCUGGGCAU .....-..--.....--((((((((....))))))))...(((((..((((((....((((.((.(.(......).).)).))))))))))..))))).. ( -23.50) >DroPse_CAF1 1728 100 + 1 AUACGAUUGGUUGAUUGCAGUUCUCCUUCGAGAGCUGCAAGUUCAAAAUACAACGCAGCAAAGAGGGCACUGGUCGUAUUGUGGUCUGGAUGCUGGGCAU (((((((..((((((((((((((((....)))))))))))..)))............((.......))))..)))))))....(((..(...)..))).. ( -33.50) >DroEre_CAF1 2022 99 + 1 GUAUC-UCACCCCAUCGCAGUUCUCCUUCGAGAGCUGCAAGUUCAAGAUCCAGCGCAGUAAGGAGGGCACCGGACGCAUCGUGGUCUGGAUGCUGGGCAU .....-..........(((((((((....)))))))))........(.((((((((..........)).((((((........))))))..))))))).. ( -34.30) >DroAna_CAF1 1947 96 + 1 AUAUC-CC---UUAUCCUAGUUCUCAUUUGAGAGCUGCAAGUUCAAGAUCCAACGAAGCAAGGAGGGCACCGGCCGCAUCGUUGUCUGGAUGCUGGGCAU ....(-((---.(((((((((((((....))))))))........(((..((((((.((......(((....))))).))))))))))))))..)))... ( -30.90) >DroPer_CAF1 1584 100 + 1 AUUCGAUUGAUUGAUUGCAGUUCUCCUUCGAGAGCUGCAAGUUCAAAAUACAACGCAGCAAAGAGGGCACUGGUCGUAUUGUGGUCUGGAUGCUGGGCAU ......((((....(((((((((((....)))))))))))..)))).......(.(((((...(((.(((..........))).)))...))))).)... ( -32.60) >consensus AUACC_UC__UCCAUUGCAGUUCUCCUUCGAGAGCUGCAAGUUCAAAAUCCAACGCAGCAAGGAGGGCACCGGACGCAUCGUGGUCUGGAUGCUGGGCAU ................(((((((((....)))))))))...............(.(((((...(((.(((..........))).)))...))))).)... (-22.78 = -22.57 + -0.22)

| Location | 13,547,121 – 13,547,217 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13547121 96 - 22224390 AUGCCCAGCAUCCAGACCACGAUACGUCCGGUGCCCUCCUUGCUGCGCUGAAUUUUGAACUUGCAGCUCUCGAAGGAGAACUGCAAUGGGUUGG-GG--- ...((((((..((((((........)))((((((..........))))))..........((((((.((((....)))).))))))))))))))-).--- ( -34.00) >DroVir_CAF1 1320 95 - 1 AUGCCCAGCAUCCAAACAACAAUGCGUCCUGUCCCUUCCUUGCUACGUUGGAUCUUAAACUUACAGCUCUCAAACGAGAACUG--AUGGA--GA-AAUAU .(((...)))((((..((((...(((..............)))...)))).............(((.((((....)))).)))--.))))--..-..... ( -16.84) >DroPse_CAF1 1728 100 - 1 AUGCCCAGCAUCCAGACCACAAUACGACCAGUGCCCUCUUUGCUGCGUUGUAUUUUGAACUUGCAGCUCUCGAAGGAGAACUGCAAUCAACCAAUCGUAU ((((.............((.((((((((((((.........)))).)))))))).))...((((((.((((....)))).))))))..........)))) ( -28.40) >DroEre_CAF1 2022 99 - 1 AUGCCCAGCAUCCAGACCACGAUGCGUCCGGUGCCCUCCUUACUGCGCUGGAUCUUGAACUUGCAGCUCUCGAAGGAGAACUGCGAUGGGGUGA-GAUAC ..((((.(((((........)))))(((((((((..........))))))))).......((((((.((((....)))).))))))..))))..-..... ( -39.30) >DroAna_CAF1 1947 96 - 1 AUGCCCAGCAUCCAGACAACGAUGCGGCCGGUGCCCUCCUUGCUUCGUUGGAUCUUGAACUUGCAGCUCUCAAAUGAGAACUAGGAUAA---GG-GAUAU ...(((.(((((.(((((((((.((((..((......)))))).))))))..))).))...)))((.((((....)))).)).......---))-).... ( -25.80) >DroPer_CAF1 1584 100 - 1 AUGCCCAGCAUCCAGACCACAAUACGACCAGUGCCCUCUUUGCUGCGUUGUAUUUUGAACUUGCAGCUCUCGAAGGAGAACUGCAAUCAAUCAAUCGAAU ((((...))))......((.((((((((((((.........)))).)))))))).))...((((((.((((....)))).)))))).............. ( -28.30) >consensus AUGCCCAGCAUCCAGACCACAAUACGUCCGGUGCCCUCCUUGCUGCGUUGGAUCUUGAACUUGCAGCUCUCGAAGGAGAACUGCAAUGAA__GA_GAUAU ..((.(((((...((..(((..........)))..))...))))).))............((((((.((((....)))).)))))).............. (-17.75 = -18.58 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:54 2006