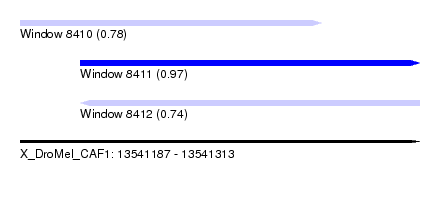
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,541,187 – 13,541,313 |
| Length | 126 |
| Max. P | 0.974013 |

| Location | 13,541,187 – 13,541,282 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13541187 95 + 22224390 GUUUUUUUUUUCAA-GGGUCAAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUGAAUCAAUUGGA ..............-.((((((........))))))..(..(((((((...((....(((((..(...---....)..)))))...)))))))))..). ( -24.70) >DroSec_CAF1 23198 91 + 1 GUUU----UUUCAG-GGGUCAAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUGAAGCAAUUGGA ((((----(.((((-.((((((........))))))..))))...............(((((..(...---....)..)))))....)))))....... ( -21.30) >DroSim_CAF1 24125 91 + 1 GUUU----UUUCAG-GGGUCAAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACCGGGGG---GAGGCGGGUGGCAAAUGAAGCAAUUGGA ((((----(.((((-.((((((........))))))..))))...............((((((.(...---....).))))))....)))))....... ( -25.30) >DroEre_CAF1 23769 90 + 1 GCUU-----UGCAG-GGGUCAAGCAAGAGAUUGGCCAACUGAUUGAUUAAAUAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUAAAUCAAUUGGA ....-----.....-.((((((........))))))..(..(((((((...((....(((((..(...---....)..)))))...)))))))))..). ( -24.20) >DroYak_CAF1 36059 94 + 1 GUUU-----UGAAGGGGGUCAAACAAGAGAUUGGCCAACUGAUUGAUUAAACAGACGGCCAGAGGGGGCGGCAGGCGGGUGGCAAAUGAAUCAAUUGGA ....-----.......((((((.(....).))))))..(..(((((((.......((.((.....)).)).((.((.....))...)))))))))..). ( -23.40) >consensus GUUU____UUUCAG_GGGUCAAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG___GAGGCGGGUGGCAAAUGAAUCAAUUGGA ................((((((........))))))..(..(((((((...((....(((((................)))))...)))))))))..). (-20.69 = -20.97 + 0.28)

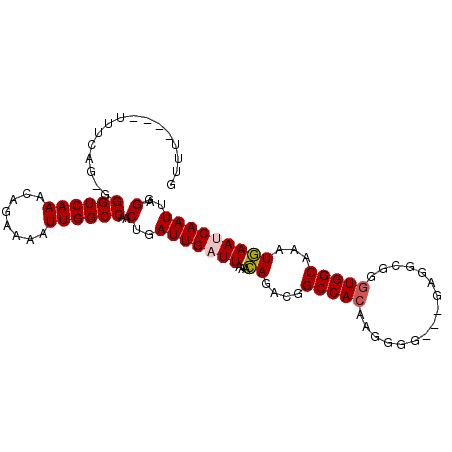

| Location | 13,541,206 – 13,541,313 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.03 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13541206 107 + 22224390 AAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUGAAUCAAUUGGAGGUCAAUUGCUGAGCAAUUUGUGGCGCAAUU ...(((..((((((((..(..(((((((...((....(((((..(...---....)..)))))...)))))))))..).)))))))).))).((........))...... ( -30.60) >DroSec_CAF1 23213 107 + 1 AAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUGAAGCAAUUGGAGGUCAAUUGCUGAGCAAUUUGUGGCGCAAUU ........(((((((((...(((((......((....(((((..(...---....)..)))))...)).((((((((.....))))))))...)))))..)))).))))) ( -31.70) >DroSim_CAF1 24140 107 + 1 AAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACCGGGGG---GAGGCGGGUGGCAAAUGAAGCAAUUGGAGGUCAAUUGCUGAGCAAUUUGUGGCGCAAUU ........(((((((((...(((((......((....((((((.(...---....).))))))...)).((((((((.....))))))))...)))))..)))).))))) ( -35.70) >DroEre_CAF1 23783 96 + 1 AAGCAAGAGAUUGGCCAACUGAUUGAUUAAAUAGACGGCCACAAGGGG---GAGGCGGGUGGCAAAUAAAUCAAUUGGAGGUCAA-----------UUUGUGGCGCAAUU ..((.(.(((((((((..(..(((((((...((....(((((..(...---....)..)))))...)))))))))..).))))))-----------))).).))...... ( -30.50) >DroYak_CAF1 36074 110 + 1 AAACAAGAGAUUGGCCAACUGAUUGAUUAAACAGACGGCCAGAGGGGGCGGCAGGCGGGUGGCAAAUGAAUCAAUUGGAGGUCAAUUGCUGAGCAAUUUGUGGCGCAAUU ..........((((((..(((.((.....)))))..))))))...(.((.(((.((...((((((.(((.((.....))..))).)))))).))....))).)).).... ( -31.00) >consensus AAACAGAAAAUUGGCCAACUGAUUGAUUAAACAGACGGCCACAAGGGG___GAGGCGGGUGGCAAAUGAAUCAAUUGGAGGUCAAUUGCUGAGCAAUUUGUGGCGCAAUU ........(((((((((.(..(((((((...((....(((((................)))))...)))))))))..).....((((((...))))))..)))).))))) (-24.03 = -24.27 + 0.24)

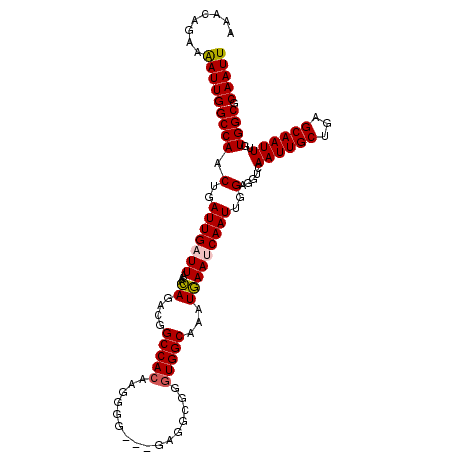
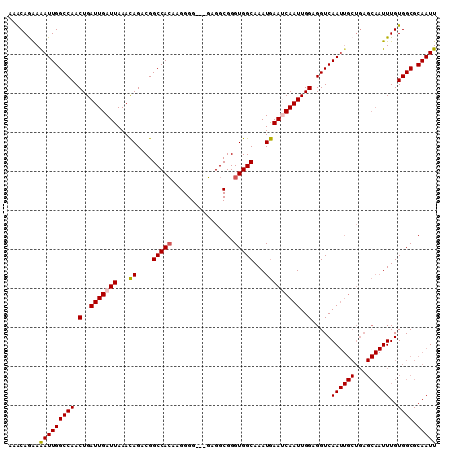
| Location | 13,541,206 – 13,541,313 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13541206 107 - 22224390 AAUUGCGCCACAAAUUGCUCAGCAAUUGACCUCCAAUUGAUUCAUUUGCCACCCGCCUC---CCCCUUGUGGCCGUCUGUUUAAUCAAUCAGUUGGCCAAUUUUCUGUUU (((((.(((...((((((...)))))).......((((((((.(((...((..((((.(---......).)).))..))...))).))))))))))))))))........ ( -21.80) >DroSec_CAF1 23213 107 - 1 AAUUGCGCCACAAAUUGCUCAGCAAUUGACCUCCAAUUGCUUCAUUUGCCACCCGCCUC---CCCCUUGUGGCCGUCUGUUUAAUCAAUCAGUUGGCCAAUUUUCUGUUU (((((.(((((..((((...((((((((.....))))))))......(((((.......---......)))))............))))..).)))))))))........ ( -23.92) >DroSim_CAF1 24140 107 - 1 AAUUGCGCCACAAAUUGCUCAGCAAUUGACCUCCAAUUGCUUCAUUUGCCACCCGCCUC---CCCCCGGUGGCCGUCUGUUUAAUCAAUCAGUUGGCCAAUUUUCUGUUU (((((.(((((..((((...((((((((.....))))))))......((((((.(....---...).))))))............))))..).)))))))))........ ( -27.60) >DroEre_CAF1 23783 96 - 1 AAUUGCGCCACAAA-----------UUGACCUCCAAUUGAUUUAUUUGCCACCCGCCUC---CCCCUUGUGGCCGUCUAUUUAAUCAAUCAGUUGGCCAAUCUCUUGCUU ......((.....(-----------(((....((((((((((.(((.(((((.......---......))))).........))).)))))))))).)))).....)).. ( -17.12) >DroYak_CAF1 36074 110 - 1 AAUUGCGCCACAAAUUGCUCAGCAAUUGACCUCCAAUUGAUUCAUUUGCCACCCGCCUGCCGCCCCCUCUGGCCGUCUGUUUAAUCAAUCAGUUGGCCAAUCUCUUGUUU .((((.(((...((((((...)))))).......((((((((.(((...((..((...((((.......))))))..))...))).)))))))))))))))......... ( -21.80) >consensus AAUUGCGCCACAAAUUGCUCAGCAAUUGACCUCCAAUUGAUUCAUUUGCCACCCGCCUC___CCCCUUGUGGCCGUCUGUUUAAUCAAUCAGUUGGCCAAUUUUCUGUUU .((((.(((...((((((...)))))).......((((((((.(((...((..((((.............)).))..))...))).)))))))))))))))......... (-16.32 = -17.12 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:52 2006