
| Sequence ID | X_DroMel_CAF1 |
|---|---|
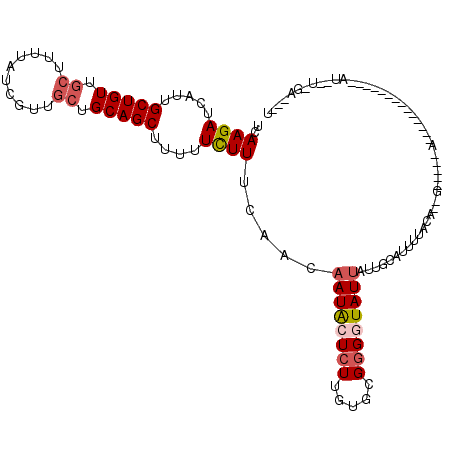
| Location | 13,532,684 – 13,532,789 |
| Length | 105 |
| Max. P | 0.877802 |

| Location | 13,532,684 – 13,532,789 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.75 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13532684 105 + 22224390 UCAAGAUCAUUGCUGUUGCUUCUAUCGUUGCUGCAGCUUUUUCUUUCAACAAUACUCUUGUGCGGGUUAUUAUUGCACUUUGCAAAG-----AACCUG---UAUCUAAU--U-GA---U ..((((.....(((((.((..........)).)))))....))))....((((......(((((((((....((((.....))))..-----))))))---)))...))--)-).---. ( -25.50) >DroSec_CAF1 14774 83 + 1 UCAAGAUCAUUGCUGUUGCUUUUAUCGUUGCUGCAGCUUUUUCUUUCAACAAUACUCUUGUGCGGGGUAU------GUAUCAA------------------------AU--U-GA---U ..((((.....(((((.((..........)).)))))....))))...(((.((((((.....)))))))------)).....------------------------..--.-..---. ( -16.80) >DroSim_CAF1 15563 88 + 1 UCAAGAUCAUUGCUGUUGCUUUUAUCGUUGCUGCAGCUUUUUCUUUC-ACAAUACUCUUGUGCGGGGUAUUGUUGCAUUUUAC------------------------AU--A-GA---U ..((((.....(((((.((..........)).)))))....))))..-((((((((((.....))))))))))..........------------------------..--.-..---. ( -20.90) >DroEre_CAF1 15057 115 + 1 UCAAGAUCAUUGCUGUUGCUUUUAUCGCUGCUGCAGCUUUUUCUUUCAAAAAUACUCUUCUGUGGGAUAUUAUUGCAUUUUAUAGCGCGAUGAUCUUAGGCUUUGUAAAGGA-AU---U ..((((((((((((((.((..........)).)))(((........(((.(((((((......))).)))).)))........))))))))))))))...............-..---. ( -25.99) >DroYak_CAF1 16822 103 + 1 UCAAGAUCAUUGCUGUUGCUUUUAUCGCU---GCAGCGUUUUUUUUCAAAAAUGCUCUUCUGUGUGGUAUUAUUGUAUUUUUUAGAGAGAUGAA-------------AAGCUAAUCUGU ...((((....(((((.((.......)).---)))))(((((((..........((((.(((...(((((....)))))...)))))))..)))-------------))))..)))).. ( -21.10) >consensus UCAAGAUCAUUGCUGUUGCUUUUAUCGUUGCUGCAGCUUUUUCUUUCAACAAUACUCUUGUGCGGGGUAUUAUUGCAUUUUACA__G_____A______________AU__U_GA___U ..((((.....(((((.((..........)).)))))....)))).....((((((((.....))))))))................................................ (-11.92 = -12.80 + 0.88)

| Location | 13,532,684 – 13,532,789 |
|---|---|
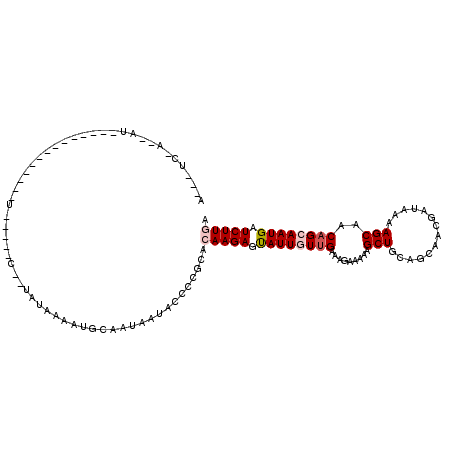
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.75 |
| Mean single sequence MFE | -18.16 |
| Consensus MFE | -7.58 |
| Energy contribution | -8.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13532684 105 - 22224390 A---UC-A--AUUAGAUA---CAGGUU-----CUUUGCAAAGUGCAAUAAUAACCCGCACAAGAGUAUUGUUGAAAGAAAAAGCUGCAGCAACGAUAGAAGCAACAGCAAUGAUCUUGA .---..-.--........---((((.(-----(.((((...((((...........)))).....((((((((..((......))....)))))))).........)))).)).)))). ( -22.20) >DroSec_CAF1 14774 83 - 1 A---UC-A--AU------------------------UUGAUAC------AUACCCCGCACAAGAGUAUUGUUGAAAGAAAAAGCUGCAGCAACGAUAAAAGCAACAGCAAUGAUCUUGA .---..-.--..------------------------.......------..........(((((.((((((((..((......))....))))))))...((....)).....))))). ( -13.60) >DroSim_CAF1 15563 88 - 1 A---UC-U--AU------------------------GUAAAAUGCAACAAUACCCCGCACAAGAGUAUUGU-GAAAGAAAAAGCUGCAGCAACGAUAAAAGCAACAGCAAUGAUCUUGA .---((-(--.(------------------------((.....)))(((((((..(......).)))))))-...)))....((((..((..........))..))))........... ( -13.30) >DroEre_CAF1 15057 115 - 1 A---AU-UCCUUUACAAAGCCUAAGAUCAUCGCGCUAUAAAAUGCAAUAAUAUCCCACAGAAGAGUAUUUUUGAAAGAAAAAGCUGCAGCAGCGAUAAAAGCAACAGCAAUGAUCUUGA .---..-..............(((((((((.((((........)).....((((.........(((.(((((....))))).)))((....)))))).........)).))))))))). ( -22.50) >DroYak_CAF1 16822 103 - 1 ACAGAUUAGCUU-------------UUCAUCUCUCUAAAAAAUACAAUAAUACCACACAGAAGAGCAUUUUUGAAAAAAAACGCUGC---AGCGAUAAAAGCAACAGCAAUGAUCUUGA ..((((((((((-------------(((...............................))))))).(((((....))))).((((.---.((.......))..))))..))))))... ( -19.18) >consensus A___UC_A__AU______________U_____C__UAUAAAAUGCAAUAAUACCCCGCACAAGAGUAUUGUUGAAAGAAAAAGCUGCAGCAACGAUAAAAGCAACAGCAAUGAUCUUGA ...........................................................(((((.((((((((.........(((..............)))..)))))))).))))). ( -7.58 = -8.42 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:48 2006