
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,487,553 – 13,487,669 |
| Length | 116 |
| Max. P | 0.885214 |

| Location | 13,487,553 – 13,487,669 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -3.14 |
| Energy contribution | -4.82 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
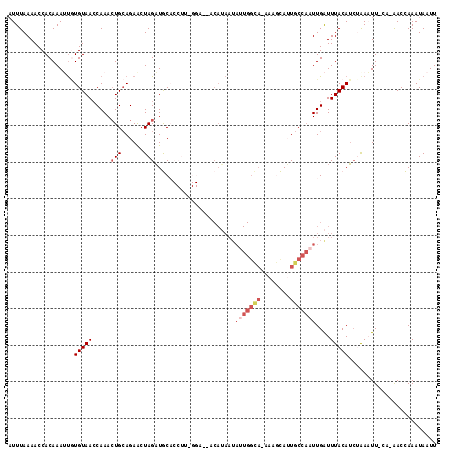
>X_DroMel_CAF1 13487553 116 + 22224390 UUUUAAAACCACAAAUUGUGUAACCAAAGUGCAGAGAUAGAUGCACCUUAGGAGCACAUAAUAUUGGCA-AAAGCAUUGCCAAUUGAUUUACAUCUGAUUUUAAUAACCAAAUAAUU ..((((((.((.....(((((..(.((.(((((........))))).)).)..)))))....(((((((-(.....))))))))...........)).))))))............. ( -23.10) >DroSec_CAF1 20020 104 + 1 AUUUAAAACCACAAAUUAUGUAACCAAACUGAAGAACUAGAUGC-----------ACAUAAUAUUGGCA-AAAGCAUUGCCAAUUGAUUUACAUCUAAAUUGCA-AACCAAAUAAUU ..................(((((.....(....)...((((((.-----------.......(((((((-(.....)))))))).......))))))..)))))-............ ( -14.96) >DroSim_CAF1 13164 104 + 1 AUUUAAAACCACAAAUUGUGUAACCAAACUGCAGAACUAGAUGC-----------ACAUAAUAUUGGCA-AAAGCAUUGCCAAUUGAUUUACAUAUAAAUUGCA-AACCAAAUAAUU ................(((((((......((((........)))-----------)......(((((((-(.....))))))))....))))))).........-............ ( -16.30) >DroEre_CAF1 3963 98 + 1 AUUUAAAACCACAAAACCUGUAACCAAACUGCAGAACUAGAUACACCUUAGGA----------------CAAAAUAAUGCCUCUUGGUUUACAUCAAAUUU-UU-AACUAAAUA-UU .....((((((......(((((.......)))))...............(((.----------------((......)))))..))))))...........-..-.........-.. ( -11.00) >DroYak_CAF1 4276 114 + 1 AUUUAAAACCACAAAAUGUGUAAACAAACUGCAGAACUAGUUACACCUUUGGAUCACAAAAUAUUGGUAAAAAAUAAUGCCACUUGGUUUACAACAAAAUU-UA-GACUAAAUA-UU .....((((((((((..(((((.....((((......))))))))).)))).............(((((........)))))..))))))...........-..-.........-.. ( -14.90) >consensus AUUUAAAACCACAAAUUGUGUAACCAAACUGCAGAACUAGAUGCACCUU_GGA__ACAUAAUAUUGGCA_AAAGCAUUGCCAAUUGAUUUACAUCUAAAUU_CA_AACCAAAUAAUU ..................(((((.....(((......)))......................(((((((........)))))))....)))))........................ ( -3.14 = -4.82 + 1.68)


| Location | 13,487,553 – 13,487,669 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -7.93 |
| Energy contribution | -8.21 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
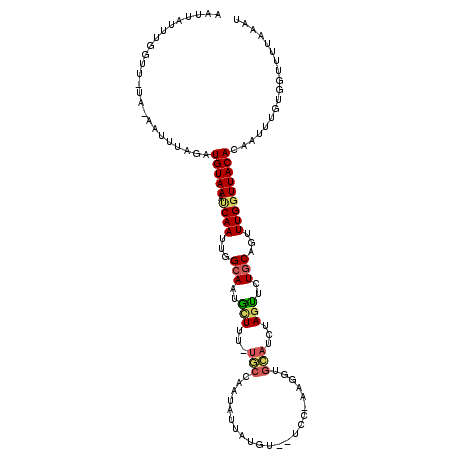
>X_DroMel_CAF1 13487553 116 - 22224390 AAUUAUUUGGUUAUUAAAAUCAGAUGUAAAUCAAUUGGCAAUGCUUU-UGCCAAUAUUAUGUGCUCCUAAGGUGCAUCUAUCUCUGCACUUUGGUUACACAAUUUGUGGUUUUAAAA ...(((((((((.....))))))))).((((((((((((((.....)-)))))))....((((...((((((((((........))))))))))...)))).....))))))..... ( -33.60) >DroSec_CAF1 20020 104 - 1 AAUUAUUUGGUU-UGCAAUUUAGAUGUAAAUCAAUUGGCAAUGCUUU-UGCCAAUAUUAUGU-----------GCAUCUAGUUCUUCAGUUUGGUUACAUAAUUUGUGGUUUUAAAU ....((((((.(-..(((.(((((((((.....((((((((.....)-)))))))......)-----------)))))))).......((......)).....)))..)..)))))) ( -22.80) >DroSim_CAF1 13164 104 - 1 AAUUAUUUGGUU-UGCAAUUUAUAUGUAAAUCAAUUGGCAAUGCUUU-UGCCAAUAUUAUGU-----------GCAUCUAGUUCUGCAGUUUGGUUACACAAUUUGUGGUUUUAAAU .......(((((-((((.......)))))))))((((((((.....)-))))))).......-----------.....(((..(..(((.(((......))).)))..)..)))... ( -24.80) >DroEre_CAF1 3963 98 - 1 AA-UAUUUAGUU-AA-AAAUUUGAUGUAAACCAAGAGGCAUUAUUUUG----------------UCCUAAGGUGUAUCUAGUUCUGCAGUUUGGUUACAGGUUUUGUGGUUUUAAAU .(-((((.(((.-..-..))).)))))((((((.(((((......(((----------------(.(((((.((((........)))).)))))..))))))))).))))))..... ( -15.00) >DroYak_CAF1 4276 114 - 1 AA-UAUUUAGUC-UA-AAUUUUGUUGUAAACCAAGUGGCAUUAUUUUUUACCAAUAUUUUGUGAUCCAAAGGUGUAACUAGUUCUGCAGUUUGUUUACACAUUUUGUGGUUUUAAAU ..-.((((((.(-((-.....(((.((((((.(((..(((..(((..((((.....(((((.....)))))..))))..)))..)))..)))))))))))).....)))..)))))) ( -18.30) >consensus AAUUAUUUGGUU_UA_AAUUUAGAUGUAAAUCAAUUGGCAAUGCUUU_UGCCAAUAUUAUGU__UCC_AAGGUGCAUCUAGUUCUGCAGUUUGGUUACACAAUUUGUGGUUUUAAAU ........................(((((.((((...(((..(((...(((......................)))...)))..)))...))))))))).................. ( -7.93 = -8.21 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:33 2006