
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,477,833 – 13,478,008 |
| Length | 175 |
| Max. P | 0.952640 |

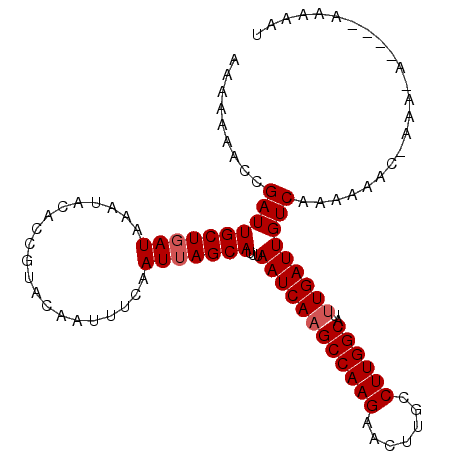
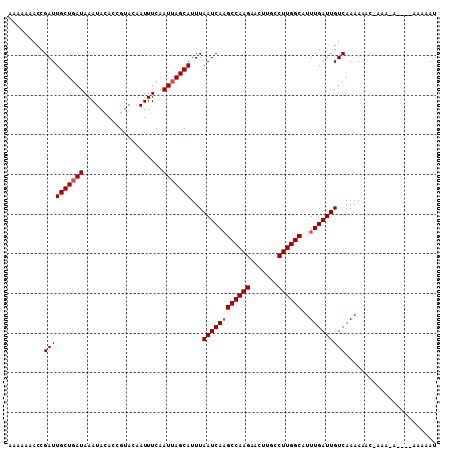
| Location | 13,477,833 – 13,477,935 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.12 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13477833 102 + 22224390 AAAAAAACCGAUUGCUGAUAAAUACACCGUACAAUUUCAAUGAGCAUUUAAUCAAGCCAAGAACUUGCCUUGGCAUUUGAUUGUCAAAAAACGAAA------AAAAAU ........((..((((.((....................)).))))..(((((((((((((.......))))))..)))))))........))...------...... ( -16.25) >DroEre_CAF1 22209 96 + 1 --AAAAACCGAUUGCUGAUAAAUAGACCGUACAAUUUCAAUUAGCAUUUAAUCAAGCCAAGAACUUGCCUUGGCAUUUGAUUGUCAAAAA------CA----AAACAU --.......((((((((((((((.(......).))))..)))))))...((((((((((((.......))))))..))))))))).....------..----...... ( -17.90) >DroYak_CAF1 22320 108 + 1 AAAAAAACCGAUUGCUGAUAAAUACACCGUACAAUUUCAAUUAGCAUUUAAUCAUGCCAAGAACUUGCCUUGGCAUUUGAUUGUCAAAAAACCAAAAAAAACAAAAAU .........((((((((((....................)))))))...((((((((((((.......)))))))..))))))))....................... ( -18.15) >consensus AAAAAAACCGAUUGCUGAUAAAUACACCGUACAAUUUCAAUUAGCAUUUAAUCAAGCCAAGAACUUGCCUUGGCAUUUGAUUGUCAAAAAAC_AAA_A____AAAAAU .........((((((((((....................)))))))...((((((((((((.......))))))..)))))))))....................... (-15.72 = -16.38 + 0.67)

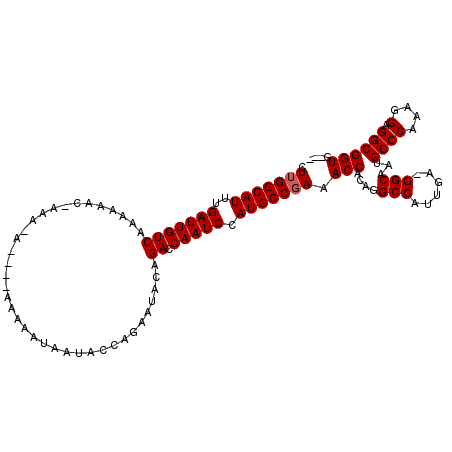
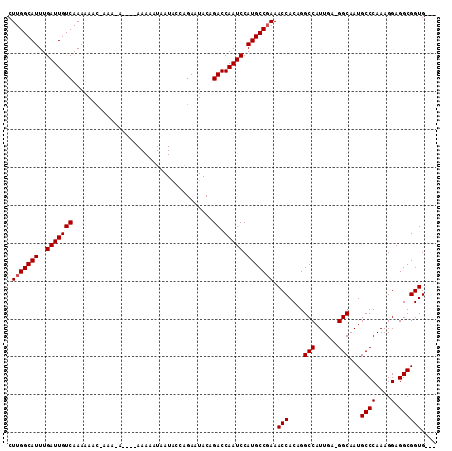
| Location | 13,477,901 – 13,478,008 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.19 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13477901 107 + 22224390 CUUGGCAUUUGAUUGUCAAAAAACGAAA------AAAAAUAAUACCAGAGUACAGACCAAUCCAUGCCGAAACCACAGGCCAUUGAAGGCAAUGCCCAAAGGAGGCGGUGGUG .(((((((..(((((((...........------....................)).))))).))))))).(((((..(((......)))..(((((....).))))))))). ( -26.98) >DroEre_CAF1 22275 99 + 1 CUUGGCAUUUGAUUGUCAAAAA------CA----AAACAUAAUGUACGACUACAGACCAAUCCAUGCCGAAACCACUGGCCAUUGA-GGCAAUGCCCAAAGGAGGCGGUG--- .(((((((..(((((((.....------..----..((.....)).........)).))))).))))))).(((.((..((.(((.-(((...)))))).)).)).))).--- ( -24.87) >DroYak_CAF1 22388 109 + 1 CUUGGCAUUUGAUUGUCAAAAAACCAAAAAAAACAAAAAUAAUACCAGAAUACAGACCAAUCCAUGCCCAAACCACAGGCCAUUGA-GGCAAUGCCCAAAGGAGGCGGUG--- ...(((((..(((((((.....................................)).))))).)))))...(((.(...((.(((.-(((...)))))).))..).))).--- ( -20.59) >consensus CUUGGCAUUUGAUUGUCAAAAAAC_AAA_A____AAAAAUAAUACCAGAAUACAGACCAAUCCAUGCCGAAACCACAGGCCAUUGA_GGCAAUGCCCAAAGGAGGCGGUG___ .(((((((..(((((((.....................................)).))))).))))))).(((....(((......)))...((((....).)))))).... (-20.86 = -21.19 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:24 2006