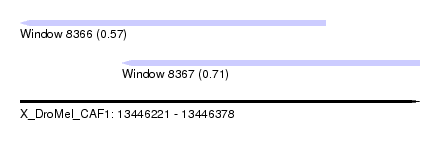
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,446,221 – 13,446,378 |
| Length | 157 |
| Max. P | 0.713710 |

| Location | 13,446,221 – 13,446,341 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.93 |
| Mean single sequence MFE | -47.98 |
| Consensus MFE | -32.66 |
| Energy contribution | -33.14 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13446221 120 - 22224390 CUGUUGGAGACCACGCUCCCGGAGAGCGGAGUGCAGCUCCUCCGCUCGACGACGAAAGACUCGUCAGGAUCGGCAGUGCUCUACGUAGAGCACCGGCGCAUUGGGAUCCAGCUCUUCGCC ..((((((..(((((.(((.(..((((((((........))))))))..)(((((.....))))).))).))((.(((((((....)))))))..))....)))..))))))........ ( -55.60) >DroPse_CAF1 12432 117 - 1 CUGCUGGAGACCGCGCUCUUGCAGAGCCGCGUGUAGCUCUUCGGCCCGACGACGGAAAACGCGUCAGGAGCGGCAGUGUUCGAUGUAGAGGACUGGCUGCGAAGGAUCCAGGGCUG---C .....(((((((((((((.....))).)))).))..)))).((((((((((.((.....)))))).(((((((((((.(((......))).))).)))))......))).))))))---. ( -48.10) >DroEre_CAF1 297 120 - 1 GUGUUGGAGCCCACGCUCCUGCAGAGCGGAGUGCAGCUCCUCCGCUCGACGACGAAAGACUCGUCAGGAUCGGCAGUGCUCUAUGUAUAGCGCCGGCUUAUUGGGAUCCAACUCUUCGGC (.((((((.((((((.((((...((((((((........))))))))((((((....)..))))))))).))((.(((((........)))))..))....)))).)))))).)...... ( -52.30) >DroWil_CAF1 12306 120 - 1 UUGCUUCAAUCCGCGUUCCUGCAAUGUGGCAUGUAUCUCGGUGGCACGUUUGCCAAAGGUAUGACAGGAGCGGCAAUGUUCAAUAUAGAGGACUGGCUUUCCUGGAUCGAGUCCCUCAUU ....(((.((((.(((((((((((((((.(((........))).))))).((((...)))))).))))))))((...((((........))))..))......)))).)))......... ( -34.30) >DroPer_CAF1 10558 117 - 1 CUGCUGGAGACCGCGCUCUUGCAGAGCCGCGUGUAGCUCUUCGGCCCGACGACGGAAAACGCGUCAGGAGCGGCAGUGUUCGAUGUAGAGGACUGGCUGCGAGGGAUCCAGGGCUG---C ((((.((((......)))).))))........((((((((....(((((((.((.....))))))....((((((((.(((......))).))).)))))..)))....)))))))---) ( -49.60) >consensus CUGCUGGAGACCGCGCUCCUGCAGAGCGGCGUGUAGCUCCUCGGCCCGACGACGAAAGACGCGUCAGGAGCGGCAGUGUUCGAUGUAGAGGACUGGCUGAGAGGGAUCCAGCGCUUC__C ..((((((..((..((((((...((((((((........))))))))((((..........))))))))))(.((((.(((......))).)))).)......)).))))))........ (-32.66 = -33.14 + 0.48)



| Location | 13,446,261 – 13,446,378 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.28 |
| Mean single sequence MFE | -50.26 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.76 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13446261 117 - 22224390 GCCGUGGUGCUCCA---AGUGCAUUCAGCUGGAGCUGAAGCUGUUGGAGACCACGCUCCCGGAGAGCGGAGUGCAGCUCCUCCGCUCGACGACGAAAGACUCGUCAGGAUCGGCAGUGCU ((((((((.(((((---(..((.((((((....))))))))..)))))))))))(.(((.(..((((((((........))))))))..)(((((.....))))).))).))))...... ( -60.70) >DroPse_CAF1 12469 117 - 1 GACGAGGCGUCCCU---UCGACAUUCAGCUGAAGCUGCAGCUGCUGGAGACCGCGCUCUUGCAGAGCCGCGUGUAGCUCUUCGGCCCGACGACGGAAAACGCGUCAGGAGCGGCAGUGUU ((((..(((..(((---..........((((((((((((((.((.(....).))((((.....)))).)).)))))))..)))))..((((.((.....)))))))))..).))..)))) ( -45.00) >DroEre_CAF1 337 117 - 1 GCCGAGGUGCUCCA---AGUGCAUUCAGCUGGAGCUGAAGGUGUUGGAGCCCACGCUCCUGCAGAGCGGAGUGCAGCUCCUCCGCUCGACGACGAAAGACUCGUCAGGAUCGGCAGUGCU (((((((.((((((---(.(((.((((((....)))))).))))))))))))....((((...((((((((........))))))))((((((....)..))))))))))))))...... ( -60.80) >DroWil_CAF1 12346 120 - 1 GUCGUGGUGGACGAUCCAGGGCAUUGAGCUGCAAUUGAAGUUGCUUCAAUCCGCGUUCCUGCAAUGUGGCAUGUAUCUCGGUGGCACGUUUGCCAAAGGUAUGACAGGAGCGGCAAUGUU (((.(((........))).))).....(((((..((((((...))))))(((.(((.(((((((((((.(((........))).)))).))))...))).)))...))))))))...... ( -39.80) >DroPer_CAF1 10595 117 - 1 GACGAGGCGUCCCU---UCGACAUUCAGCUGAAGCUGCAGCUGCUGGAGACCGCGCUCUUGCAGAGCCGCGUGUAGCUCUUCGGCCCGACGACGGAAAACGCGUCAGGAGCGGCAGUGUU ((((..(((..(((---..........((((((((((((((.((.(....).))((((.....)))).)).)))))))..)))))..((((.((.....)))))))))..).))..)))) ( -45.00) >consensus GACGAGGUGCCCCA___AGGGCAUUCAGCUGAAGCUGAAGCUGCUGGAGACCGCGCUCCUGCAGAGCGGCGUGUAGCUCCUCGGCCCGACGACGAAAGACGCGUCAGGAGCGGCAGUGUU ....................((.((((((....))))))(((((.(....)..(((((((...((((((((........))))))))((((..........)))))))))))))))))). (-28.40 = -28.76 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:12 2006