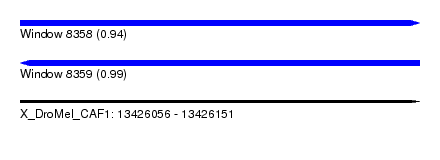
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,426,056 – 13,426,151 |
| Length | 95 |
| Max. P | 0.986431 |

| Location | 13,426,056 – 13,426,151 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -12.23 |
| Energy contribution | -13.28 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
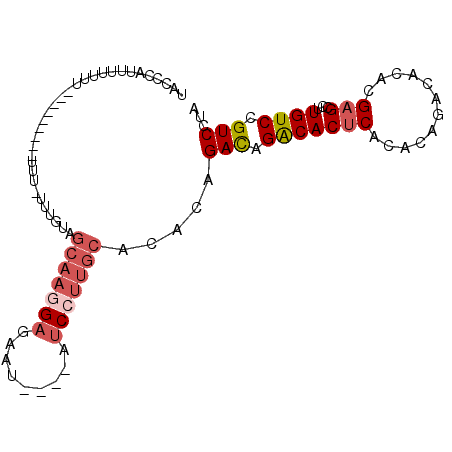
>X_DroMel_CAF1 13426056 95 + 22224390 UAUCCAUUUUUUUUUUU-UUUUUUUUGUUGUGGCAAGGUGAAU----AUCCUUGCACACAGACAGACACUCACACAGACACACGAGCCUUGUCCGUCCUA .................-..........((((((((((.....----..)))))).))))(((.(((((((............)))...)))).)))... ( -21.30) >DroSec_CAF1 21587 96 + 1 UACCCAUUUUUUUUUUUGUUUUUUUUUUUGUAGCAAGGAGAAU----AUCCUUGCACACAGACAGACACUCACACAGACACACGAGCCUUGUCCGUCCUA ............................(((.(((((((....----.))))))))))..(((.(((((((............)))...)))).)))... ( -20.00) >DroSim_CAF1 20888 87 + 1 UACCCAUUUUUUU---------UUUUUUUGUAGCAAGGAGAAU----AUCCUUGCACACAGACAGACACUCACACAGACACACGAGCCUUGUCCGUCCUA .............---------......(((.(((((((....----.))))))))))..(((.(((((((............)))...)))).)))... ( -20.00) >DroEre_CAF1 16504 79 + 1 UAGCCAUUUUUUU-------------A----CGCAAAGAGGAU----AUCGUUGCACACAGAUAGACACCCACACAGACACACGAGCCUUGUCCGUCCUA .............-------------.----.((((.((....----.)).)))).....((..((((.....................))))..))... ( -8.70) >DroYak_CAF1 31263 83 + 1 UAACCUUUUUUUU-------------UUUGUAGCAAGGAGUAU----AUCCUUGCACACAGACAGACACUCACACAGACACACGAGCCUUGUCCGUCCUA .............-------------..(((.(((((((....----.))))))))))..(((.(((((((............)))...)))).)))... ( -20.00) >DroPer_CAF1 33607 89 + 1 ----CAUUAU----UUUGUUUUCUU-UUUG--CCACUGAACGCACUCCUCCACACACACAGACAGGCACUCACACAGCCAUAUGAGCCUUGUCCGUCCUA ----......----..(((.(((..-....--.....))).))).............((.((((((..((((..........)))).)))))).)).... ( -10.90) >consensus UACCCAUUUUUUU_________UUU_UUUGUAGCAAGGAGAAU____AUCCUUGCACACAGACAGACACUCACACAGACACACGAGCCUUGUCCGUCCUA ................................(((((((.........))))))).....(((.(((((((............)))...)))).)))... (-12.23 = -13.28 + 1.06)


| Location | 13,426,056 – 13,426,151 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
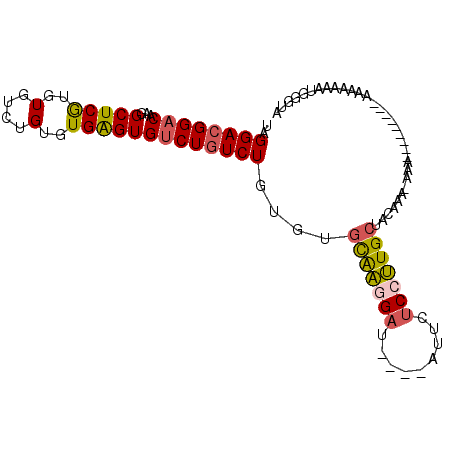
>X_DroMel_CAF1 13426056 95 - 22224390 UAGGACGGACAAGGCUCGUGUGUCUGUGUGAGUGUCUGUCUGUGUGCAAGGAU----AUUCACCUUGCCACAACAAAAAAAA-AAAAAAAAAAAUGGAUA ..((((((((...(((((..(....)..)))))))))))))(((.((((((..----.....)))))))))...........-................. ( -29.70) >DroSec_CAF1 21587 96 - 1 UAGGACGGACAAGGCUCGUGUGUCUGUGUGAGUGUCUGUCUGUGUGCAAGGAU----AUUCUCCUUGCUACAAAAAAAAAAACAAAAAAAAAAAUGGGUA ..((((((((...(((((..(....)..)))))))))))))(((.(((((((.----....))))))))))............................. ( -29.30) >DroSim_CAF1 20888 87 - 1 UAGGACGGACAAGGCUCGUGUGUCUGUGUGAGUGUCUGUCUGUGUGCAAGGAU----AUUCUCCUUGCUACAAAAAAA---------AAAAAAAUGGGUA ..((((((((...(((((..(....)..)))))))))))))(((.(((((((.----....)))))))))).......---------............. ( -29.30) >DroEre_CAF1 16504 79 - 1 UAGGACGGACAAGGCUCGUGUGUCUGUGUGGGUGUCUAUCUGUGUGCAACGAU----AUCCUCUUUGCG----U-------------AAAAAAAUGGCUA ..(((..(((...(((((..(....)..))))))))..)))....((((.((.----....)).)))).----.-------------............. ( -15.60) >DroYak_CAF1 31263 83 - 1 UAGGACGGACAAGGCUCGUGUGUCUGUGUGAGUGUCUGUCUGUGUGCAAGGAU----AUACUCCUUGCUACAAA-------------AAAAAAAAGGUUA ..((((((((...(((((..(....)..)))))))))))))(((.(((((((.----....))))))))))...-------------............. ( -29.30) >DroPer_CAF1 33607 89 - 1 UAGGACGGACAAGGCUCAUAUGGCUGUGUGAGUGCCUGUCUGUGUGUGUGGAGGAGUGCGUUCAGUGG--CAAA-AAGAAAACAAA----AUAAUG---- ..(.(((((((.(((((((((....))))))..)))))))))).).(((.......(((........)--))..-......)))..----......---- ( -22.06) >consensus UAGGACGGACAAGGCUCGUGUGUCUGUGUGAGUGUCUGUCUGUGUGCAAGGAU____AUUCUCCUUGCUACAAA_AAA_________AAAAAAAUGGGUA ..((((((((...(((((..(....)..)))))))))))))....(((((((.........)))))))................................ (-20.31 = -20.48 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:04 2006