
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,420,945 – 13,421,084 |
| Length | 139 |
| Max. P | 0.996618 |

| Location | 13,420,945 – 13,421,054 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.97 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -8.73 |
| Energy contribution | -10.46 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
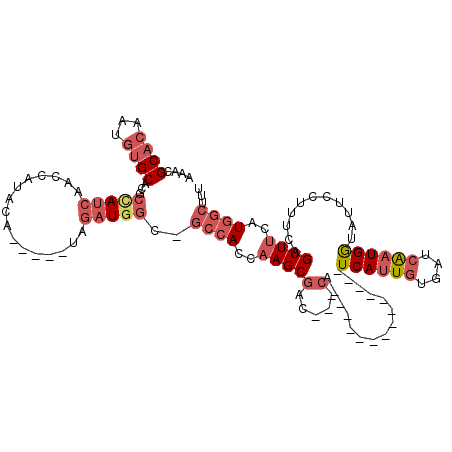
>X_DroMel_CAF1 13420945 109 - 22224390 AACCGCACAAUGUGCACGCCAUCAACCAUACA-----UAGAUGGC-GCCACCAAGCGAC---CAUCAAACCCACAUCAUCAUUGUGAUCAAUGAUAUUCCUUUCAGUUUCAUGGCUUC ....((((...))))..(((((.(((......-----..((((((-((......))).)---))))...........(((((((....)))))))..........)))..)))))... ( -29.40) >DroGri_CAF1 6986 96 - 1 GCAAGCUAAACGUGCCCCAAACCAACCAAAUGCAUCUUAUAUCCCAGGCAUGGAGCCAA---CG--------------UCAUUAUCAACG-UGGUUAUCAAUAUGGUUGCUU----GU ((((((....((((((..............................)))))).(((((.---((--------------(........)))-)))))............))))----)) ( -20.51) >DroSec_CAF1 16569 95 - 1 AAACGCACAAUGUGCACGCCAUCAACCAUACA-----UAGAUGGC-GCCACCAAGCGAC---CA--------------UCAUUGUGAUCAAUGGUAUUCCUUUCAGUUUCAUGGCUUU ....((((...))))..(((((.(((..((((-----..((((((-((......))).)---))--------------))..))))......((....)).....)))..)))))... ( -25.80) >DroSim_CAF1 15843 95 - 1 AAACGCACAAUGUGCACGCCAUCAACCAUACA-----UAGAUGGC-GCCACCAAGCGAC---CA--------------UCAUUGUGAUCAAUGGUAUUCCUUACAGUUUCAUGGCUUU ....((((...))))..((((((.........-----..))))))-((((..((((...---..--------------.(((((....)))))(((.....))).))))..))))... ( -26.50) >DroEre_CAF1 11370 101 - 1 AAGCGCACAAAGUGCGUACUAUC--------A-----CAGAUGGC-GCCACCAAGCGAC---CAUCAAACCCACAUCAUCAUUGUGAUCAAUGGUAUUCCUUUCAGUUUUAUGGCUUU ((((((((...))))(((((((.--------.-----..((((((-((......))).)---)))).....((((.......))))....)))))))................)))). ( -26.20) >DroYak_CAF1 26216 108 - 1 AAGCGCACAAUGUGCAUACUAUCAACC----A-----UAGAUGGC-GCCAGCAAGCGAACCACAUCAAACCCACAUCAUCAUUGUGAUCAAUGGUGUUCCUUUCAGUUUCAUGGCUUU ..((((.....))))...(((((....----.-----..))))).-((((..(((((((..((((((....((((.......)))).....))))))....))).))))..))))... ( -25.10) >consensus AAACGCACAAUGUGCACGCCAUCAACCAUACA_____UAGAUGGC_GCCACCAAGCGAC___CA______________UCAUUGUGAUCAAUGGUAUUCCUUUCAGUUUCAUGGCUUU ....((((...))))...(((((................)))))..((((..(((((.....)...............((((((....))))))...........))))..))))... ( -8.73 = -10.46 + 1.72)

| Location | 13,420,985 – 13,421,084 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.90 |
| Mean single sequence MFE | -17.34 |
| Consensus MFE | -11.98 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
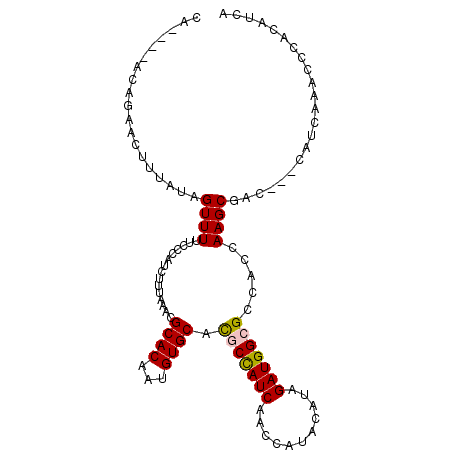
>X_DroMel_CAF1 13420985 99 - 22224390 CA----ACAGAACUUUAUAGUUUUUCCCAUUUUUAACCGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC---CAUCAAACCCACAUCA ..----..((((((....))))))..............((((...))))...................((((((((......))).)---))))............ ( -18.00) >DroSec_CAF1 16609 84 - 1 CA----AGAGA-UUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC---CA-------------- ..----(((((-(...............))))))...(((.....(((..(((((((...........))))))).)))...)))..---..-------------- ( -16.96) >DroSim_CAF1 15883 84 - 1 CA----AGAGA-UUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC---CA-------------- ..----(((((-(...............))))))...(((.....(((..(((((((...........))))))).)))...)))..---..-------------- ( -16.96) >DroEre_CAF1 11410 94 - 1 AAAAAUGCAGAACAUCAAAGUUUUUC-CAUCUAUAAGCGCACAAAGUGCGUACUAUC--------ACAGAUGGCGCCACCAAGCGAC---CAUCAAACCCACAUCA ....(((.(((((......)))))..-)))......((((((...))))))......--------...((((((((......))).)---))))............ ( -20.70) >DroYak_CAF1 26256 102 - 1 AAAGAAGCAGAACAUCAUAGUUUUUCCCAACAUUAAGCGCACAAUGUGCAUACUAUCAACC----AUAGAUGGCGCCAGCAAGCGAACCACAUCAAACCCACAUCA ......((....((((...(((......))).....((((.....))))............----...))))((....))..))...................... ( -14.10) >consensus CA____ACAGAACUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC___CAUCAAACCCACAUCA ...................((((...............((((...)))).(((((((...........))))))).....))))...................... (-11.98 = -11.90 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:02 2006