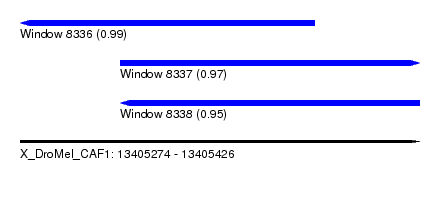
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,405,274 – 13,405,426 |
| Length | 152 |
| Max. P | 0.994437 |

| Location | 13,405,274 – 13,405,386 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13405274 112 - 22224390 AAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAGUUUUGUUUGAC-- (((((....(((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..))))).(((((((((.(.....).)))).))))))))))...-- ( -28.10) >DroSec_CAF1 1166 112 - 1 AAACAUUUUUUCAUUUGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGGCAGUUUGGUUUGAC-- ((((.....(((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..)))))((((((((((.(.....).)))).))))))))))...-- ( -30.90) >DroSim_CAF1 1166 112 - 1 AAACAUUCUUUCAUUUGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAGUUUGGUUUGAC-- ((((.....(((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..)))))((((((((((.(.....).)))).))))))))))...-- ( -31.40) >DroYak_CAF1 8983 120 - 1 AAACAUUCCUUUAUUCACACAUUUUGCUUGGCAUUCUCGAACUACAUAUUUAAUAAAUGGUAAAUAAAAUGUGUCUGUUAGCAAAUUUCUCUGAUUUCCUGAGACAGUUUUGUUUGACAA ................((((((((((.(((.((((....................)))).))).)))))))))).((((((((((..((((.........))))....)))).)))))). ( -20.55) >consensus AAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU__CGAACU____AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAGUUUGGUUUGAC__ ((((.....(((((..(((((.((((((((.((((..((((.......))))...)))).)))))))).)))))..)))))((((((((((.(.....).)))).))))))))))..... (-26.08 = -26.57 + 0.50)


| Location | 13,405,312 – 13,405,426 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13405312 114 + 22224390 UCACAGACACAUUUUGUUUGCCAUUUAUCAAAU----AGUUCG--AAUGCCAAACAAAGUGUGCGAAUGAAAGAAUGUUUGCCAAUGCGAAAGAACUUAAAGCCUUUCAAAUAUUUAUAA (((..(.(((((((((((((.(((((.......----.....)--)))).))))))))))))))...)))..((((((((((....))(((((..........))))))))))))).... ( -27.00) >DroSec_CAF1 1204 114 + 1 UCACAGACACAUUUUGUUUGCCAUUUAUCAAAU----AGUUCG--AAUGCCAAACAAAGUGUGCAAAUGAAAAAAUGUUUGCCAAAGCAAAAGAACUUAAAGCCUUUCAAAUAUUUAUAA (((....(((((((((((((.(((((.......----.....)--)))).)))))))))))))....)))..(((((((((.....((.............))....))))))))).... ( -23.72) >DroSim_CAF1 1204 114 + 1 UCACAGACACAUUUUGUUUGCCAUUUAUCAAAU----AGUUCG--AAUGCCAAACAAAGUGUGCAAAUGAAAGAAUGUUUGCCAAAGCAAAAGAACUUAAAGCCUUUCAAAUAUUUAUAA .....(.(((((((((((((.(((((.......----.....)--)))).))))))))))))))...((((((..(.(((((....))))).........)..))))))........... ( -24.90) >DroYak_CAF1 9023 119 + 1 UAACAGACACAUUUUAUUUACCAUUUAUUAAAUAUGUAGUUCGAGAAUGCCAAGCAAAAUGUGUGAAUAAAGGAAUGUUUGC-AAAGCUAAAGAACUUAAAGCCUUUCAAAUAUUUAUAA ......(((((((((.(((..(((((.(..(((.....)))..))))))..))).)))))))))........(((((((((.-((((((...........)).))))))))))))).... ( -19.10) >consensus UCACAGACACAUUUUGUUUGCCAUUUAUCAAAU____AGUUCG__AAUGCCAAACAAAGUGUGCAAAUGAAAGAAUGUUUGCCAAAGCAAAAGAACUUAAAGCCUUUCAAAUAUUUAUAA .....(.(((((((((((((.((((...(.(((.....))).)..)))).))))))))))))))........(((((((((.....((.............))....))))))))).... (-20.01 = -19.57 + -0.44)


| Location | 13,405,312 – 13,405,426 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13405312 114 - 22224390 UUAUAAAUAUUUGAAAGGCUUUAAGUUCUUUCGCAUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGA ............(((((((.....).))))))((....))..........((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..)))) ( -27.30) >DroSec_CAF1 1204 114 - 1 UUAUAAAUAUUUGAAAGGCUUUAAGUUCUUUUGCUUUGGCAAACAUUUUUUCAUUUGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGA ....((((.((((.(((((.............)))))..)))).))))..((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..)))) ( -28.02) >DroSim_CAF1 1204 114 - 1 UUAUAAAUAUUUGAAAGGCUUUAAGUUCUUUUGCUUUGGCAAACAUUCUUUCAUUUGCACACUUUGUUUGGCAUU--CGAACU----AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGA .................(((..((((......)))).)))..........((((..(((((.((((((((.((((--(((...----..))))...))).)))))))).)))))..)))) ( -27.50) >DroYak_CAF1 9023 119 - 1 UUAUAAAUAUUUGAAAGGCUUUAAGUUCUUUAGCUUU-GCAAACAUUCCUUUAUUCACACAUUUUGCUUGGCAUUCUCGAACUACAUAUUUAAUAAAUGGUAAAUAAAAUGUGUCUGUUA .........((((.((((((...........))))))-.)))).............((((((((((.(((.((((....................)))).))).))))))))))...... ( -17.95) >consensus UUAUAAAUAUUUGAAAGGCUUUAAGUUCUUUUGCUUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU__CGAACU____AUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGA .........((((.(((((.............)))))..)))).......((((..(((((.((((((((.((((..((((.......))))...)))).)))))))).)))))..)))) (-22.28 = -22.72 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:46 2006