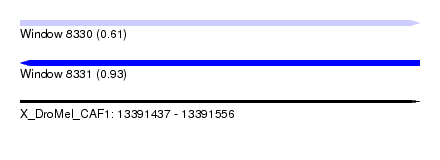
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,391,437 – 13,391,556 |
| Length | 119 |
| Max. P | 0.932526 |

| Location | 13,391,437 – 13,391,556 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13391437 119 + 22224390 AGUGUCAACGAGAAGGAGUACACGGUGCGGGCGGAUGUCAAGGACACGCCACCCACCUCCGAUUUUGACGACGAGCUGAGUGGUCAGUUCUAAAAAUUGGUCAAAGGGAUUUCCCUUUA .(((((..(.....)..).))))((((.(((((..((((...)))))))).).)))).((((((((......(((((((....)))))))..))))))))..((((((....)))))). ( -39.60) >DroSec_CAF1 4030 119 + 1 AGUGUCAACGAGAAGGAGUACACUGUGCGGGCGGAUGUCAAGGACACGCCACCCUCCUCCGAUUUCGACGACGAGCUGAGUGGUCAACUUUAAAAAUUACUUAAAGGGAAAUGCCUUUA ..((((..(((((.(((((((...))))(((.((.((((...))))..)).)))...)))..)))))..))))...((((((((...........))))))))(((((.....))))). ( -31.20) >DroSim_CAF1 7454 119 + 1 AGUGUCAACGAGAAGGAGUACACGGUGCGGGCGGAUGUCAAGGACACGCCACCCACCUCCGAUUUCGACGACGAGCUGAGUGGUCAGCUCUAAAAAUGAGUCAAAGGUAAAUGCCUUUA ...(((..(((((.((((....)((((.(((((..((((...)))))))).).)))))))..)))))..)))(((((((....)))))))............((((((....)))))). ( -40.90) >DroEre_CAF1 6867 112 + 1 AGUGUCAACGAGAAGGAGUACACGGUGCGGGCGGAUGUCAAGGACACGCCACCCACCUCCGAUUUCGAUGACGAGCUGAAUGGUCAGCUCUAGGAAUUGGGCAA-------UCCCUGCC ...((((.(((((.((((....)((((.(((((..((((...)))))))).).)))))))..))))).))))(((((((....)))))))..((....(((...-------.)))..)) ( -41.30) >DroYak_CAF1 4423 107 + 1 AGUGUCAACGAGAAGGAGUACACGGUGCGUGCGGAUGUUAAGGACACGCCACCCAGCUCCGAUUUCGAGGACGAGCUUAAUGGUCAGCUCUAGAAGUUGGCCAA-------GAA----- ..((((..(((((.(((((....(((((((((..........).)))).))))..)))))..)))))..)))).......(((((((((.....))))))))).-------...----- ( -38.50) >DroAna_CAF1 4394 100 + 1 AAUGUCAACGACAAGGACUACACGGUGCGGGCCGAUGUGAAGGACACAGCGCCCACCUCUGACUUCGAUGAUGAGCUGAGUGGUCAGCUUUGAA--UCAGGU----------------- ...((((.......(.(((....))).)((((...((((.....))))..)))).....))))...(((...(((((((....)))))))...)--))....----------------- ( -29.50) >consensus AGUGUCAACGAGAAGGAGUACACGGUGCGGGCGGAUGUCAAGGACACGCCACCCACCUCCGAUUUCGACGACGAGCUGAGUGGUCAGCUCUAAAAAUUAGUCAA_______UCCCUUUA ...(((..(((((.(((((((...))))(((.((.((((...))))..)).)))...)))..)))))..)))(((((((....)))))))............................. (-25.62 = -26.23 + 0.61)

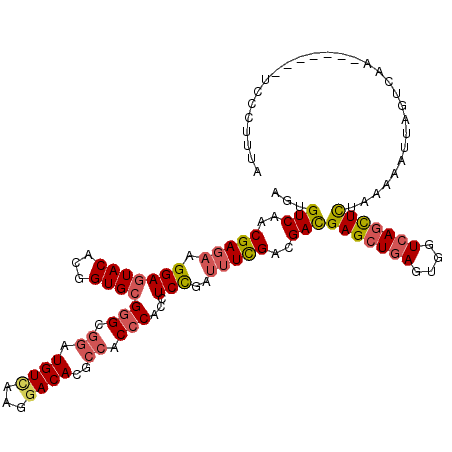

| Location | 13,391,437 – 13,391,556 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -27.71 |
| Energy contribution | -29.93 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
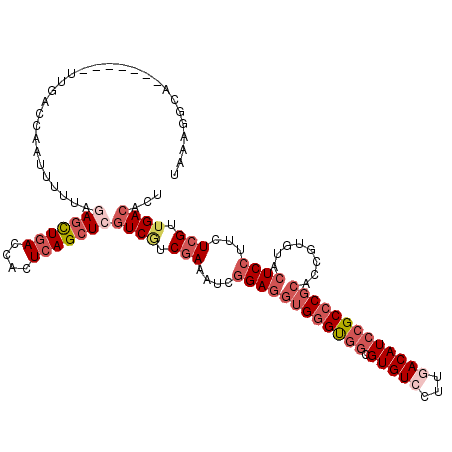
>X_DroMel_CAF1 13391437 119 - 22224390 UAAAGGGAAAUCCCUUUGACCAAUUUUUAGAACUGACCACUCAGCUCGUCGUCAAAAUCGGAGGUGGGUGGCGUGUCCUUGACAUCCGCCCGCACCGUGUACUCCUUCUCGUUGACACU (((((((....)))))))...........((.((((....)))).))...(((((....((((((((((((.(((((...)))))))))))))((...)).))))......)))))... ( -40.70) >DroSec_CAF1 4030 119 - 1 UAAAGGCAUUUCCCUUUAAGUAAUUUUUAAAGUUGACCACUCAGCUCGUCGUCGAAAUCGGAGGAGGGUGGCGUGUCCUUGACAUCCGCCCGCACAGUGUACUCCUUCUCGUUGACACU ((((((......))))))............((((((....)))))).((((.(((....((((((((((((.(((((...)))))))))))((.....))..))))))))).))))... ( -37.30) >DroSim_CAF1 7454 119 - 1 UAAAGGCAUUUACCUUUGACUCAUUUUUAGAGCUGACCACUCAGCUCGUCGUCGAAAUCGGAGGUGGGUGGCGUGUCCUUGACAUCCGCCCGCACCGUGUACUCCUUCUCGUUGACACU ((((((......))))))...........(((((((....)))))))((((.(((....((((((((((((.(((((...)))))))))))))((...)).))))...))).))))... ( -45.00) >DroEre_CAF1 6867 112 - 1 GGCAGGGA-------UUGCCCAAUUCCUAGAGCUGACCAUUCAGCUCGUCAUCGAAAUCGGAGGUGGGUGGCGUGUCCUUGACAUCCGCCCGCACCGUGUACUCCUUCUCGUUGACACU ...(((((-------((....))))))).(((((((....)))))))((((.(((....((((((((((((.(((((...)))))))))))))((...)).))))...))).))))... ( -47.90) >DroYak_CAF1 4423 107 - 1 -----UUC-------UUGGCCAACUUCUAGAGCUGACCAUUAAGCUCGUCCUCGAAAUCGGAGCUGGGUGGCGUGUCCUUAACAUCCGCACGCACCGUGUACUCCUUCUCGUUGACACU -----...-------..............(((((........)))))(((..(((....((((...((((.(((((...........))))))))).....))))...)))..)))... ( -29.90) >DroAna_CAF1 4394 100 - 1 -----------------ACCUGA--UUCAAAGCUGACCACUCAGCUCAUCAUCGAAGUCAGAGGUGGGCGCUGUGUCCUUCACAUCGGCCCGCACCGUGUAGUCCUUGUCGUUGACAUU -----------------..((((--((....(((((....)))))((......)))))))).((((((.(((((((.....))).)))))).))))..........((((...)))).. ( -27.40) >consensus UAAAGGCA_______UUGACCAAUUUUUAGAGCUGACCACUCAGCUCGUCGUCGAAAUCGGAGGUGGGUGGCGUGUCCUUGACAUCCGCCCGCACCGUGUACUCCUUCUCGUUGACACU .............................(((((((....)))))))((((.(((....((((((((((((.(((((...)))))))))))))........))))...))).))))... (-27.71 = -29.93 + 2.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:39 2006