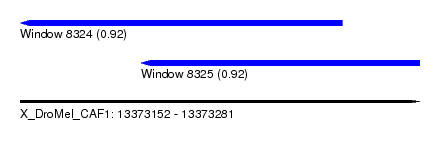
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,373,152 – 13,373,281 |
| Length | 129 |
| Max. P | 0.924491 |

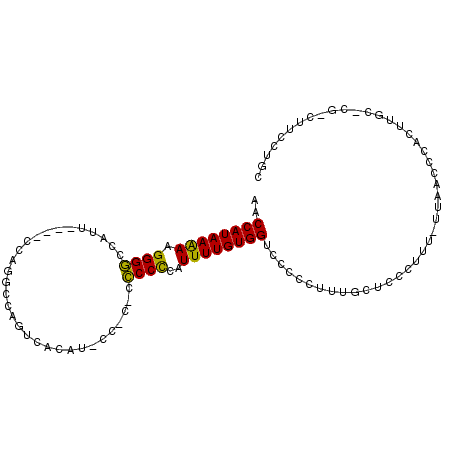
| Location | 13,373,152 – 13,373,256 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -17.03 |
| Consensus MFE | -11.73 |
| Energy contribution | -10.85 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13373152 104 - 22224390 AACCAUAAAAAGGGGCCAUUCCUUCCAGGCCAGUCACAUCCCUCCCCCCUCAUUUUGUGGUCCCCCUUUGCUCCCUUUGCUAACCCACUUGCCCGACUUCCUGC .((((((((((((((((..........))))((........))....)))..)))))))))........((.......))........................ ( -16.00) >DroSim_CAF1 6147 90 - 1 AACCAUAAAAAGGGGCCAUU----CCAGGCCAGUCAUAUACCCCACCCCCCAUUUUGUGGGCCCCCUUUGCUCCCUUU-UUAACCCA---------CUUCCUGC .....((((((((((((...----...))))..........(((((..........)))))............)))))-))).....---------........ ( -21.60) >DroEre_CAF1 8615 88 - 1 AACCAUAAGAGGGGUCAGU-----CCGGUCCAGUCACA--------GCCCAAUUUUGUGGUUCCCUUUC-CUUCCUUU-UUAACCCACUUGC-CGUCUUCCUGC ((((((((((.((((....-----.(......).....--------))))..)))))))))).......-........-.............-........... ( -13.50) >consensus AACCAUAAAAAGGGGCCAUU____CCAGGCCAGUCACAU_CC_C_CCCCCCAUUUUGUGGUCCCCCUUUGCUCCCUUU_UUAACCCACUUGC_CG_CUUCCUGC ..((((((((.((((...............................))))..))))))))............................................ (-11.73 = -10.85 + -0.88)


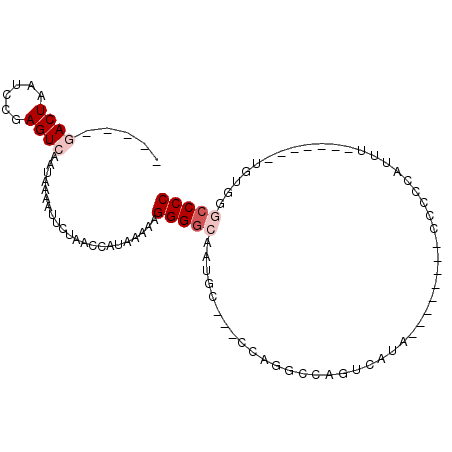
| Location | 13,373,191 – 13,373,281 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 68.28 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -5.04 |
| Energy contribution | -6.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.25 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13373191 90 - 22224390 -----GACUAAUCCGAGUCAAUAAAAUUCUAACCAUAAAAAGGGGCCAUUCCUUCCAGGCCAGUCACAUCCCUCCCCCCUCAUUU-------UGUGGUCCCC -----((((......))))............((((((((((((((((..........))))((........))....)))..)))-------)))))).... ( -18.90) >DroPse_CAF1 11134 86 - 1 -----AACUAAUCCAAGUCAAUAAAAUGCUAAACGUAAAAAGGGGCAU-GC-A-CCAAGGCAUGCAUG--------CCGCCAUUCCACUGCCCGUUGGCCCC -----...........((((((....(((.....)))....(((((((-((-(-........))))))--------)).))............))))))... ( -21.90) >DroSim_CAF1 6176 86 - 1 -----GACUAAUCCGAGUCAAUAAAAUUCUAACCAUAAAAAGGGGCCAUU----CCAGGCCAGUCAUAUACCCCACCCCCCAUUU-------UGUGGGCCCC -----((((......)))).............((((((((.((((((...----...)))).((.....))........)).)))-------)))))..... ( -20.50) >DroEre_CAF1 8651 77 - 1 -----GACUAAUCCGAGUCAAUAAAAUUCUAACCAUAAGAGGGGUCAGU-----CCGGUCCAGUCACA--------GCCCAAUUU-------UGUGGUUCCC -----((((......))))...........((((((((((.((((....-----.(......).....--------))))..)))-------)))))))... ( -17.30) >DroPer_CAF1 11228 91 - 1 CACAAAACUAAUCCAAGUCAAUAAAAUGCUAAACGUAAAAAGGGGCAU-GC-A-CCAAGGCAUGCAUG--------CCGCCAUUCCACUGCCCGUUGGCCCC ................((((((....(((.....)))....(((((((-((-(-........))))))--------)).))............))))))... ( -21.90) >consensus _____GACUAAUCCGAGUCAAUAAAAUUCUAACCAUAAAAAGGGGCAAUGC___CCAGGCCAGUCAUA________CCCCCAUUU_______UGUGGGCCCC .....((((......))))......................(((((...................................................))))) ( -5.04 = -6.04 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:34 2006