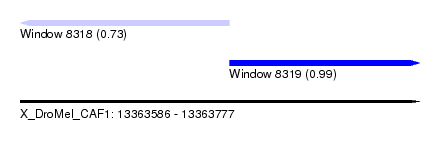
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,363,586 – 13,363,777 |
| Length | 191 |
| Max. P | 0.993496 |

| Location | 13,363,586 – 13,363,686 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13363586 100 - 22224390 AAAGACAAUAAAAACAAAUAAAAAGUAUUUUAAAAUGCCAUUUGAAUUAAACUUGAAACCGAGAACC---AACCGAAGGC----AACU--GAAACACAGUGGCAAAAAU ...................................(((((((((.......((((....))))....---......((..----..))--....)).)))))))..... ( -13.90) >DroSec_CAF1 9298 98 - 1 AAAGACAAUAAAAACAAAUAAAAAGUAUUUUGAAAUGCCAUUUGAAUUAAACUUGAAACCAAGAACC---AACCGAAGGC----AACU--GAAA--CAGUGGCAAAAAU ..............((((..........))))...(((((((.(.......((((....))))....---......((..----..))--....--))))))))..... ( -13.40) >DroSim_CAF1 9315 98 - 1 AAAGACAAUAAAAACAAAUAAAAAGUAUUUUGAAAUGCCAUUUGAAUUAAACUUGAAACCGAGAACC---AACCGAAGGC----AACU--GAAA--CAGUGGCAAAAAU ..............((((..........))))...(((((((.(.......((((....))))....---......((..----..))--....--))))))))..... ( -13.10) >DroYak_CAF1 11054 107 - 1 AAAGACAAUAAAAACAAAUAAAAAGUAUUUCGAAAUGCCAUUUGAAUUAAACUUGAAACCGAGAACUGAGAACUGAGAACCAAAAACUGAGAAA--CAGUGGCGAAAAU ...................................((((............((((....)))).((((....((.((.........)).))...--))))))))..... ( -11.60) >consensus AAAGACAAUAAAAACAAAUAAAAAGUAUUUUGAAAUGCCAUUUGAAUUAAACUUGAAACCGAGAACC___AACCGAAGGC____AACU__GAAA__CAGUGGCAAAAAU ...................................(((((((.........((((....))))..........(....)..................)))))))..... (-10.12 = -9.38 + -0.75)


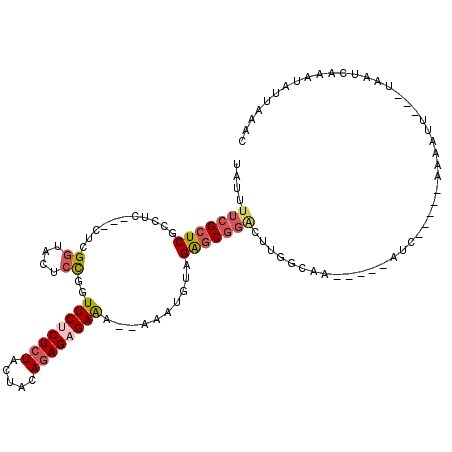
| Location | 13,363,686 – 13,363,777 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.48 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -11.75 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13363686 91 + 22224390 UAUUUUCGCUCGCCUC---CUCGGUACUCCAGUUCGCUCUACUACACAGAGAGA--AACUGUAGAUC-------ACUA-----AUC-----ACAGUU---UCAUUAAAUCUUUAAC ........((((((..---...))).....(((.......))).....))).((--((((((.(((.-------....-----)))-----))))))---)).............. ( -16.00) >DroSec_CAF1 9396 103 + 1 UAUUUUCGCUCGCCUC---CUCGGUACUCCGGUUCUCUCUGCUACAGAGAGAAAAAAAAUGUAGAGCGGACUUGGCAA-----AUC-----UAUAUUUACUAAUCAAAUAUUAAGC ....(((((((.....---.((((....))))(((((((((...)))))))))..........))))))).((((.((-----((.-----...)))).))))............. ( -25.60) >DroSim_CAF1 9413 103 + 1 UAUUUUCGCUCGCCUC---CGCGCUACUCCGGUUCCCUCUGCUACAGAGAGAAAAAAAAUGUAGGGCGGACUUGGCAA-----AUC-----UAUAUUUACUAAUCAAAUAUUAAGC .......(((.(((((---(((.((((.....(((.(((((...))))).))).......)))).)))))...)))..-----...-----.((((((.......))))))..))) ( -27.80) >DroYak_CAF1 11161 111 + 1 UAUUUUCGCUCGACUCCUUCUCGGUGCUCUUAUUCUCUCUACUACAGAGAGAGA--AAAUGUUGAGCGGGCUUGUUAAUAUUAAUGUUAAUACAGCU---CCAUUAAAACAUCAAU .......(((((((.((.....))........((((((((.......)))))))--)...)))))))(((((((((((((....)))))))).))).---)).............. ( -30.90) >consensus UAUUUUCGCUCGCCUC___CUCGGUACUCCGGUUCUCUCUACUACAGAGAGAAA__AAAUGUAGAGCGGACUUGGCAA_____AUC_____AAAAUU___UAAUCAAAUAUUAAAC ....(((((((...........((....))..((((((((.....))))))))..........))))))).............................................. (-11.75 = -13.00 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:29 2006