
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,358,377 – 13,358,481 |
| Length | 104 |
| Max. P | 0.832597 |

| Location | 13,358,377 – 13,358,481 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -32.60 |
| Energy contribution | -33.80 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13358377 104 + 22224390 ---GAGCUGUUCCAGUUGUUGUAGCCGGCGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCUGCUCAGAAAACAAUGCG----A ---((((.((.((.((((((((((((((((...)))...(((((((((...))))))).)).....))))))))))))))).)).....)))).............----. ( -41.00) >DroSec_CAF1 3849 103 + 1 ---GAGCUGUUCCAGUUGUUGUAGCCGGCGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCAGCU-AGAAAACAAUGCG----C ---.(((((..((.((((((((((((((((...)))...(((((((((...))))))).)).....)))))))))))))))......)))))-.............----. ( -41.10) >DroSim_CAF1 3828 103 + 1 ---GAGCUGUUCCAGUUGUUGUAGCCGGCGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCAGCU-AGAAAACAAUGAG----A ---.(((((..((.((((((((((((((((...)))...(((((((((...))))))).)).....)))))))))))))))......)))))-.............----. ( -41.10) >DroEre_CAF1 3425 93 + 1 -------------AGUUGUUGUAGCCGGCGGAGCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCAGCU-GGAAAACAAUGCG----A -------------.((((((((((((((((...)))...(((((((((...))))))).)).....))))))))))))).((((((((....-.)))...))))).----. ( -38.50) >DroYak_CAF1 5714 106 + 1 UGCGAGCUGUUCCAGUUGUCGUAGCCGACGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCAGCU-GGAAAACAAUGCG----A .(((.....((((.(((((((((((((..(((((((....)).(((((...)))))))....))).)))))))))))))(((.......)))-)))).....))).----. ( -41.90) >DroAna_CAF1 4052 100 + 1 ---AGCCUGUUCCAGUUGUU---GCCGCCGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUU----GCAGAGACAGCGGCAUUUUCUGGU-CGAAAACAAUGCGAGAGA ---.((.((((((((....(---(((((.(..(.((((.(((((((((...))))))).)).))----)).)...).))))))....)))).-....))))..))...... ( -35.10) >consensus ___GAGCUGUUCCAGUUGUUGUAGCCGGCGGACCGCAAUUGCUGGCGACAUUUGCCGGACAUUCUACGGCUGCGACAGCGGCAUUUUCAGCU_AGAAAACAAUGCG____A ..............((((((((((((((((...)))...(((((((((...))))))).)).....))))))))))))).((((((((......)))...)))))...... (-32.60 = -33.80 + 1.20)

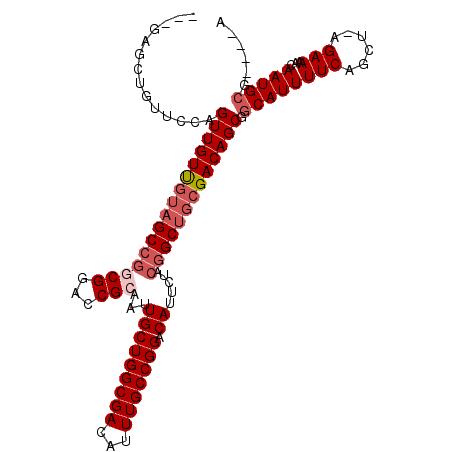
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:25 2006