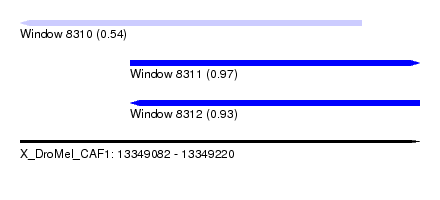
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,349,082 – 13,349,220 |
| Length | 138 |
| Max. P | 0.968510 |

| Location | 13,349,082 – 13,349,200 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13349082 118 - 22224390 CUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGGCAUCUCAAACCGGCGGCGAAUCUGCAUUUGAUGCUCACAGCUUUCAUUGUUUGAUUGGCAACCAUUUGUGAA--AUAUUC ...........(((((((((((..((.((..(((((((((.........))))))).))...)).))..))))))))))).((((((.....(((.(....).))).)))))--)..... ( -34.50) >DroSec_CAF1 10913 118 - 1 CUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCGGCGUAUCUGCAUUUGAUGCUCACAGCUUUCAUUGUUUGAUUGGCAACCAUUUGUGAA--AUAUUC ...........(((((((((((.((((.((.....)).))))..........((((.....))))....))))))))))).((((((.....(((.(....).))).)))))--)..... ( -31.10) >DroSim_CAF1 13494 114 - 1 CUAU-GAC-UCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACAG--GGCGAAUCUGCAUUUGAUGCUCACAGCUUUCAUUGUUUGAUUGGCAACCAUUUGUGAA--AUAUUC ....-...-..(((((((((((..........(((((.((....))..))))--)(((....)))....))))))))))).((((((.....(((.(....).))).)))))--)..... ( -29.80) >DroEre_CAF1 9964 118 - 1 CUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCAGCGAAUCUGCAUUUGAUGCUCGCAGCUUCCAUUGUUUGAUUGGCAGCCAUUUGUGAA--AUAUUC ...((.((....((((((((((..((.((..((((((((...........)))))).))...)).))..))))))))))(((.(((.........))).))).....)).))--...... ( -31.00) >DroYak_CAF1 16288 118 - 1 CUAUUGACUUCCUGUGAGCAUCCUGUUCACAUCCUGUCGACAUCUCAAACCGGCGGCGAAUCUGCAUUUGAUGCUCACAGCUUCCAUUGUUUGAUUGGCAACCAUUUGUGAA--AUAUUC ...(((((....(((((((.....)))))))....)))))..((.((((..((..((.((((.(((...((.((.....)).))...)))..)))).))..)).)))).)).--...... ( -32.30) >DroAna_CAF1 12049 97 - 1 -GGAUGCCUUC------ACAUCCUUU--GCAUCCUGAUAGCGCUUCU--------GAUAUUCUGCUGUUGAUGC------CUUUCAUUGUUUGAUUGGCAACCGUUUGUGAACCACAUUC -(((((.....------.)))))((.--.((..(.(((((((.....--------.......)))))))..(((------(..(((.....)))..))))...)..))..))........ ( -23.70) >consensus CUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCGGCGAAUCUGCAUUUGAUGCUCACAGCUUUCAUUGUUUGAUUGGCAACCAUUUGUGAA__AUAUUC ...........(((((((((((.((((.((.....)).))))..........((((.....))))....)))))))))))......(((((.....)))))................... (-19.84 = -20.77 + 0.92)



| Location | 13,349,120 – 13,349,220 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13349120 100 + 22224390 CUGUGAGCAUCAAAUGCAGAUUCGCCGCCGGUUUGAGAUGCCGACAGGAUGUGGACAGGAUGCUCACAGGAAGUCAAUAGACAUAUACACCAUACCCCAA (((((((((((...(.((.((((...(.((((.......)))).).)))).)).)...)))))))))))...(((....))).................. ( -32.60) >DroSec_CAF1 10951 99 + 1 CUGUGAGCAUCAAAUGCAGAUACGCCGCCGGUUUGAGAUGUCGACAGGAUGUGGACAGGAUGCUCACAGGAAGUCAAUAGACAUAUACACCAUGCCCC-A (((((((((((.....(((((.((....)))))))...((((.((.....)).)))).)))))))))))...(((....)))................-. ( -31.40) >DroSim_CAF1 13532 94 + 1 CUGUGAGCAUCAAAUGCAGAUUCGCC--CUGUUUGAGAUGUCGACAGGAUGUGGACAGGAUGCUCACAGGA-GUC-AUAGACAUAUACACCAUAC-CC-A (((((((((((...((....((((((--(((((.((....))))))))..))))))).)))))))))))..-(((-...))).............-..-. ( -30.40) >DroEre_CAF1 10002 100 + 1 CUGCGAGCAUCAAAUGCAGAUUCGCUGCCGGUUUGAGAUGUCGACAGGAUGUGGACAGGAUGCUCACAGGAAGUCAAUAGAUAUACACCACAUAUAUCAA (((.(((((((....((((.....))))..........((((.((.....)).)))).))))))).)))..........((((((.......)))))).. ( -30.60) >consensus CUGUGAGCAUCAAAUGCAGAUUCGCCGCCGGUUUGAGAUGUCGACAGGAUGUGGACAGGAUGCUCACAGGAAGUCAAUAGACAUAUACACCAUACCCC_A (((((((((((.....(((((.........)))))...((((.((.....)).)))).)))))))))))...(((....))).................. (-26.94 = -27.25 + 0.31)

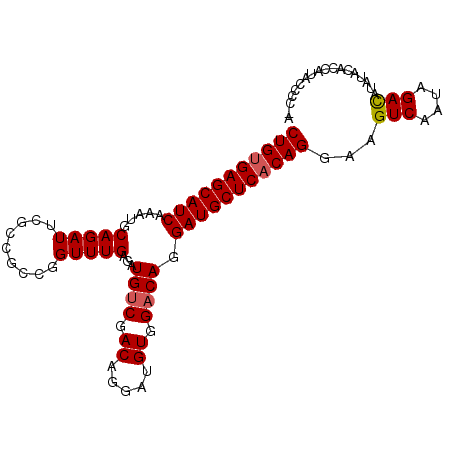
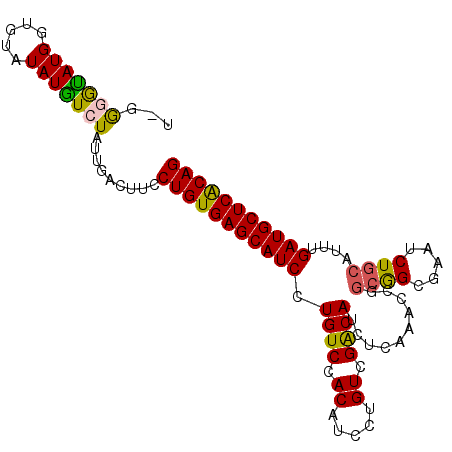
| Location | 13,349,120 – 13,349,220 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -25.96 |
| Energy contribution | -25.65 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13349120 100 - 22224390 UUGGGGUAUGGUGUAUAUGUCUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGGCAUCUCAAACCGGCGGCGAAUCUGCAUUUGAUGCUCACAG ..(((((((((.........))))..)))))(((((((((((..((.((..(((((((((.........))))))).))...)).))..))))))))))) ( -32.20) >DroSec_CAF1 10951 99 - 1 U-GGGGCAUGGUGUAUAUGUCUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCGGCGUAUCUGCAUUUGAUGCUCACAG .-.(((((((.....))))))).........(((((((((((.((((.((.....)).))))..........((((.....))))....))))))))))) ( -32.40) >DroSim_CAF1 13532 94 - 1 U-GG-GUAUGGUGUAUAUGUCUAU-GAC-UCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACAG--GGCGAAUCUGCAUUUGAUGCUCACAG .-((-((((((.........))))-.))-))(((((((((((..........(((((.((....))..))))--)(((....)))....))))))))))) ( -28.30) >DroEre_CAF1 10002 100 - 1 UUGAUAUAUGUGGUGUAUAUCUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCAGCGAAUCUGCAUUUGAUGCUCGCAG ..(((((((.....)))))))..........(((((((((((..((.((..((((((((...........)))))).))...)).))..))))))))))) ( -31.70) >consensus U_GGGGUAUGGUGUAUAUGUCUAUUGACUUCCUGUGAGCAUCCUGUCCACAUCCUGUCGACAUCUCAAACCGGCGGCGAAUCUGCAUUUGAUGCUCACAG ...(((((((.....))))))).........(((((((((((.((((.((.....)).))))..........((((.....))))....))))))))))) (-25.96 = -25.65 + -0.31)

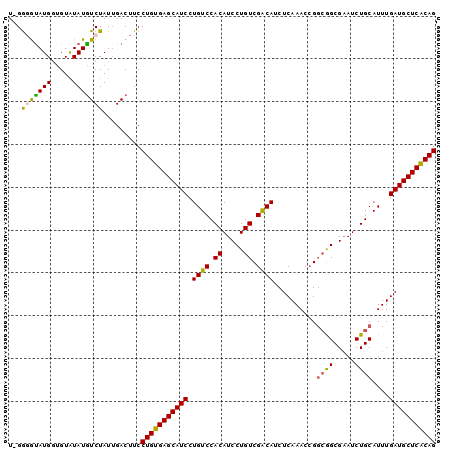
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:23 2006