| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,324,331 – 13,324,429 |
| Length | 98 |
| Max. P | 0.962364 |

| Location | 13,324,331 – 13,324,429 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.04 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
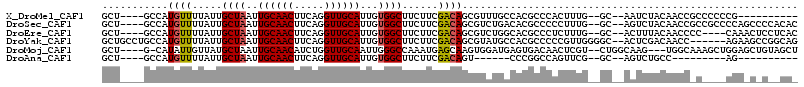
>X_DroMel_CAF1 13324331 98 + 22224390 GCU----GCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGCGUUUGCCACGCCCACUUUG--GC--AAUCUACAACCGCCCCCCG---------- ...----((..(((...((((((((.((((((.....))))))..(((((.....((....))...))))).......)))--))--)))..)))...))......---------- ( -25.80) >DroSec_CAF1 36908 108 + 1 GCU----GCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGCGUCUGACACGCCCCCUUUG--GC--AGUCUACAACCGCCGCCCCAGCCCCACAC (((----((((((((.....((((..((((((.....))))))...))))......))))((((.....))))......))--))--))).......................... ( -28.90) >DroEre_CAF1 28455 104 + 1 GCU----GCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGCGUCUGGCACGCCCUCUUUG--GC--ACUUUACAACCCC----CAAACUCCUCAC ..(----((((((((.....((((..((((((.....))))))...))))......))))((((.....))))......))--))--)............----............ ( -24.80) >DroYak_CAF1 28251 108 + 1 GCUGCCUGCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGCGUAUGCCACGCCCCCGUUGGGGC--ACUCGACAACC------AGAAGCCGGCAG .....(((((........((((.....))))((((.((((((...(((((.....((....))...)))))(((((....)))))--....).)))))------.))))..))))) ( -41.80) >DroMoj_CAF1 2887 106 + 1 GCU----G-CAUAUUGUUAUGCUAAUUGCAACAUCUGGUUGCAAUUGGGCCAAAUGAGCAAGUGGAUGAGUGACAACUCGU--CUGGCAAG---UGGCAAAGCUGGAGCUGUAGCU (((----(-(...(((((((.(((((((((((.....)))))))))))(((..((......))((((((((....))))))--)))))..)---))))))(((....)))))))). ( -41.30) >DroAna_CAF1 13362 83 + 1 GCU----GCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGU------CCCGGCCAGUUCG--GC--AGUCUGCC---------AG---------- (((----(((.((((.....((((..((((((.....))))))...))))......))))((------....))......)--))--))).....---------..---------- ( -24.60) >consensus GCU____GCCAUGUUUUAUUGCUAAUUGCAACUUCAGGUUGCAUUGUGGCUUCUUCGACAGCGUCUGCCACGCCCACUUUG__GC__AAUCUACAACC_C____CGA_C_C__CA_ ...........((((.....((((..((((((.....))))))...))))......))))........................................................ (-13.21 = -13.27 + 0.06)
| Location | 13,324,331 – 13,324,429 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.04 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -9.02 |
| Energy contribution | -11.02 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13324331 98 - 22224390 ----------CGGGGGGCGGUUGUAGAUU--GC--CAAAGUGGGCGUGGCAAACGCUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGC----AGC ----------..((.((((.((((....(--((--(......))))..)))).)))).))......((((...((((((.....)))))).(((....)))....))))----... ( -30.30) >DroSec_CAF1 36908 108 - 1 GUGUGGGGCUGGGGCGGCGGUUGUAGACU--GC--CAAAGGGGGCGUGUCAGACGCUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGC----AGC ((((...(((((((((((((((...))))--))--)...((.(((((.....))))).))......)))....((((((.....)))))).)))))......))))...----... ( -37.40) >DroEre_CAF1 28455 104 - 1 GUGAGGAGUUUG----GGGGUUGUAAAGU--GC--CAAAGAGGGCGUGCCAGACGCUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGC----AGC ((.(...((((.----.((((((((..((--((--....((.(((((.....))))).))......).)))..))))))))...(((((.....))))).)))).).))----... ( -28.60) >DroYak_CAF1 28251 108 - 1 CUGCCGGCUUCU------GGUUGUCGAGU--GCCCCAACGGGGGCGUGGCAUACGCUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGCAGGCAGC (((((((((((.------((((((.....--(((((....)))))(((((.....((.....))..)))))...)))))).)))))(((.....)))........))))))..... ( -44.30) >DroMoj_CAF1 2887 106 - 1 AGCUACAGCUCCAGCUUUGCCA---CUUGCCAG--ACGAGUUGUCACUCAUCCACUUGCUCAUUUGGCCCAAUUGCAACCAGAUGUUGCAAUUAGCAUAACAAUAUG-C----AGC .((...(((....)))..))..---...(((((--(.((((.((.........))..)))).))))))..(((((((((.....))))))))).(((((....))))-)----... ( -28.80) >DroAna_CAF1 13362 83 - 1 ----------CU---------GGCAGACU--GC--CGAACUGGCCGGG------ACUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGC----AGC ----------.(---------(((((.((--((--(.....)).))).------.)))))).....((((...((((((.....)))))).(((....)))....))))----... ( -23.80) >consensus _UG__G_G_UCG____G_GGUUGUAGACU__GC__CAAAGUGGGCGUGCCAGACGCUGUCGAAGAAGCCACAAUGCAACCUGAAGUUGCAAUUAGCAAUAAAACAUGGC____AGC ..........................................(((((.....))))).........((((...((((((.....)))))).(((....)))....))))....... ( -9.02 = -11.02 + 2.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:11 2006