
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,319,670 – 13,319,797 |
| Length | 127 |
| Max. P | 0.784338 |

| Location | 13,319,670 – 13,319,767 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13319670 97 - 22224390 CUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU-GGGGCUUAUCCCCAUACCCCUCCCCACCCGCCAC-----ACGAUUUGC-----------CGCCCAGCCGCA ...........((((((((.........(((((((.....))-))))).(((....))).............)))))-----)))...(((-----------.((...)).))) ( -26.90) >DroSec_CAF1 32292 89 - 1 CUUGAUUGCCACGUGUGGUAAUUUCCUCGUCCCGAUAAAUUU-GGGCCUUAUCCCCAC-----UCCCUUCCCACCGA-----ACGAUUUG--------------CCCAGCCGCA ...((((((((....)))))))).................((-((((...(((.....-----..............-----..)))..)--------------)))))..... ( -15.60) >DroSim_CAF1 25038 89 - 1 CUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU-GGGGCUUAUCCCCAC-----UCCCUUCCCACCGA-----GCGAUUUG--------------CCCAGCCGCA ...((((((((....)))))))).....(((((((.....))-)))))..........-----..............-----(((.....--------------......))). ( -23.50) >DroEre_CAF1 23714 106 - 1 CUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUUUGGGGCUCAUUCCCC--------CGAUUCCACCGAUUCCACCGAAUCGCCCAGCCCCCAUCCCCCAGCCUCA ...((((((((....)))))))).....(((((((......))))))).........--------.........((((((....))))))........................ ( -26.30) >consensus CUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU_GGGGCUUAUCCCCAC_____UCCCUUCCCACCGA_____ACGAUUUG______________CCCAGCCGCA ...((((((((....)))))))).....(((((.(......).))))).................................................................. (-15.38 = -15.62 + 0.25)



| Location | 13,319,693 – 13,319,797 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13319693 104 - 22224390 UUGCAA-ACAUUCCAUAGCCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU--GGGGCUUAUCCCCAUACCCCUCCCCACCCGCC ..((..-.((.((...(((.((.....)).))).)).))....((((((..((......(((((((.....))--)))))..))..))))))............)). ( -24.30) >DroSec_CAF1 32312 99 - 1 UUGCAA-ACAUUCCAUAUCCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGUCCCGAUAAAUUU--GGGCCUUAUCCCCAC-----UCCCUUCCCACC ..((((-(.((((.......)))).)))..))..((((((((....)))))))).....(.(((((.....))--))).)..........-----............ ( -14.60) >DroSim_CAF1 25058 99 - 1 UUGCAA-ACAUUCCAUAGCCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU--GGGGCUUAUCCCCAC-----UCCCUUCCCACC ......-.........(((.((.....)).))).((((((((....)))))))).....(((((((.....))--)))))..........-----............ ( -22.90) >DroEre_CAF1 23753 97 - 1 UUGCAA-GCAUUCCAUAGCCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU-UGGGGCUCAUUCCCC--------CGAUUCCACC ...(((-((((((.......))))......)))))(((((((....)))))))......(((((((......)-)))))).........--------.......... ( -24.30) >DroYak_CAF1 23464 86 - 1 CUGCAAAACAUUCCAUAGUCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGCACCGAUAAAUUUGUGGGCCUUAUUC--------------------- ....................(((((..(((((..((((((((....))))))))...(((...))).......)))))...)))))--------------------- ( -15.50) >consensus UUGCAA_ACAUUCCAUAGCCGAAUAUUUCCGCUUGAUUGCCACGUGUGGUAAUUUCCUCGCCCCGAUAAAUUU__GGGGCUUAUCCCCA______UCCCUUCCCACC ................(((.((.....)).))).((((((((....)))))))).....(((((...........)))))........................... (-13.68 = -14.32 + 0.64)

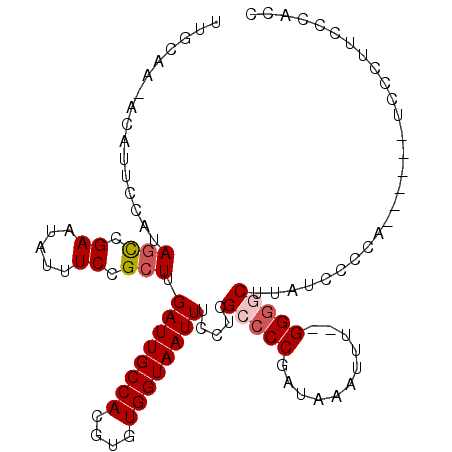
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:07 2006