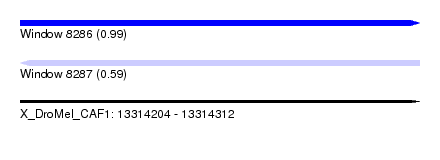
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,314,204 – 13,314,312 |
| Length | 108 |
| Max. P | 0.990920 |

| Location | 13,314,204 – 13,314,312 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.23 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13314204 108 + 22224390 UCGGGCAGGAAUGGCAAA---------ACGGCAAACGACAGCGGGGAAUUUCCCUUCUGUGUUUUUCCUCGCUUU-UUGUCUUUCCCCCCCCACUUCCAUUUUCCUCUUUUUUUCCCA ..(((..((((.((((((---------(.(((((((.((((.((((.....)))).))))))))......)))))-))))).))))..)))........................... ( -31.20) >DroSim_CAF1 19199 105 + 1 UCGGGCAGGAAUGGCAAA---------ACGGCAAACGACAGCGGGGAAUUUCCCGCCUGUGUUUUUCCUCGCUUU-UUGUCUUUCC---CCCACUUCCAUUUUCCUCUUUUUUCCCCA ..(((..((((.((((((---------(.(((((((.(((((((((....))))).))))))))......)))))-))))).))))---))).......................... ( -31.00) >DroEre_CAF1 18501 105 + 1 UCGGGCAGGAAUGGCAAA----------GGGCAAACGACAGCGGGGAAUUUCCCUCCUGUGUUUUUCCUCGCUUU-UUGUCUUUCCC--CCCACUUCCAUUUUCUUCUUUUUCCCCAC ..(((..((((.((((((----------((((((((.((((.((((.....)))).))))))))......)))))-))))).)))).--))).......................... ( -34.30) >DroYak_CAF1 17985 114 + 1 UCGGGCAGGAAUGGCAAAAGUGGCAAAACGGCAAACGACAGCGGGGAAUUUCCCUCCUGUGUU-UUCCUCGCUUUUUUGUCUUUCC---CCCACUUCCAUUUUCCCUUUUUUCCCCCG ..(((..((((.((((((((.(((.....((.((((.((((.((((.....)))).)))))))-).))..))))))))))).))))---))).......................... ( -34.20) >consensus UCGGGCAGGAAUGGCAAA_________ACGGCAAACGACAGCGGGGAAUUUCCCUCCUGUGUUUUUCCUCGCUUU_UUGUCUUUCC___CCCACUUCCAUUUUCCUCUUUUUCCCCCA ..(((..((((.(((((...........((.....))..(((((((((...(.(....).)...)))))))))...))))).))))...))).......................... (-26.35 = -26.60 + 0.25)


| Location | 13,314,204 – 13,314,312 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.23 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13314204 108 - 22224390 UGGGAAAAAAAGAGGAAAAUGGAAGUGGGGGGGGAAAGACAA-AAAGCGAGGAAAAACACAGAAGGGAAAUUCCCCGCUGUCGUUUGCCGU---------UUUGCCAUUCCUGCCCGA ...........................(((.(((((.(.(((-((.....((..((((((((..(((.....)))..)))).)))).)).)---------)))).).))))).))).. ( -30.70) >DroSim_CAF1 19199 105 - 1 UGGGGAAAAAAGAGGAAAAUGGAAGUGGG---GGAAAGACAA-AAAGCGAGGAAAAACACAGGCGGGAAAUUCCCCGCUGUCGUUUGCCGU---------UUUGCCAUUCCUGCCCGA ..(((.......((((..((((....(.(---(.((((((..-.......(......)...((((((......))))))))).))).)).)---------....)))))))).))).. ( -29.80) >DroEre_CAF1 18501 105 - 1 GUGGGGAAAAAGAAGAAAAUGGAAGUGGG--GGGAAAGACAA-AAAGCGAGGAAAAACACAGGAGGGAAAUUCCCCGCUGUCGUUUGCCC----------UUUGCCAUUCCUGCCCGA ...(((....((.....(((((....(((--(.(((.((((.-...(((.((((.....(....).....)))).))))))).))).)))----------)...))))).)).))).. ( -29.00) >DroYak_CAF1 17985 114 - 1 CGGGGGAAAAAAGGGAAAAUGGAAGUGGG---GGAAAGACAAAAAAGCGAGGAA-AACACAGGAGGGAAAUUCCCCGCUGUCGUUUGCCGUUUUGCCACUUUUGCCAUUCCUGCCCGA ...(((.....(((((...(..((((((.---((((..............((.(-(((((((..(((.....)))..)))).)))).)).)))).))))))..)...))))).))).. ( -35.16) >consensus UGGGGAAAAAAGAGGAAAAUGGAAGUGGG___GGAAAGACAA_AAAGCGAGGAAAAACACAGGAGGGAAAUUCCCCGCUGUCGUUUGCCGU_________UUUGCCAUUCCUGCCCGA .........................((((...((((.(.(((........((...(((((((..(((.....)))..)))).)))..))............))).).))))..)))). (-21.10 = -21.10 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:01 2006