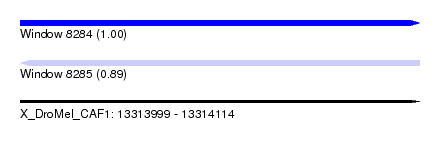
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,313,999 – 13,314,114 |
| Length | 115 |
| Max. P | 0.998762 |

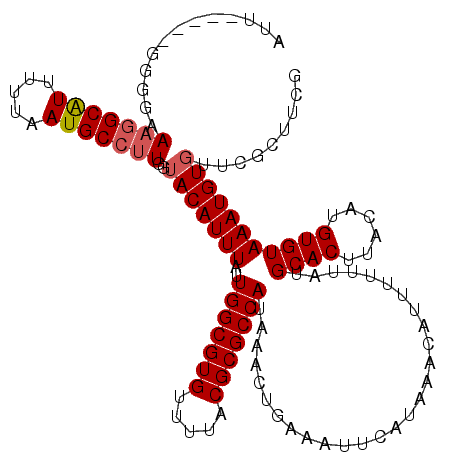
| Location | 13,313,999 – 13,314,114 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13313999 115 + 22224390 CGAAGCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAGGGCAUUAAAAAUGCCUUUCCCC-----AAU .((((......(((((((....)))))))((((((.....))))))..)))).((((((((((.(.....).)))).)))))).....(((((((.....))))))).....-----... ( -26.90) >DroVir_CAF1 64752 109 + 1 CGGAGGAGACACAUUUACACAUGUA-GUGCAUAA--AAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUUCCAAGGCAUUUAA-AUGCUUUUGCUU-------U .((((....)(((((((....((.(-((.(((((--.....))))).))).)).....(((((.(.....).))))).))))))))))(((((((....-))))))).....-------. ( -25.30) >DroPse_CAF1 74838 120 + 1 AGAACCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAACGCCUUGCUCCUCGCUACU .((((.(((..(((((((....)))))))((((((.....))))))...))).)))).(((((.(.....).)))))..........((((((.........))))))............ ( -27.40) >DroEre_CAF1 18305 115 + 1 CGAAGCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUUUCCCC-----AAU .((((......(((((((....)))))))((((((.....))))))..)))).((((((((((.(.....).)))).)))))).....(((((((.....))))))).....-----... ( -27.20) >DroYak_CAF1 17770 115 + 1 CGAAGCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUUUCCCC-----AAU .((((......(((((((....)))))))((((((.....))))))..)))).((((((((((.(.....).)))).)))))).....(((((((.....))))))).....-----... ( -27.20) >DroPer_CAF1 74767 120 + 1 AGAACCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAACGCCUUUCUCCUCGCUACU .((((.(((..(((((((....)))))))((((((.....))))))...))).)))).(((((.(.....).)))))...........(((((.........)))))............. ( -24.50) >consensus CGAAGCGAACACAUUUACACAUGUAAGUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUUUCCCC_____AAU .(((.......(((((((....)))))))((((((.....))))))...))).((((((((((.(.....).)))).)))))).....(((((((.....)))))))............. (-24.36 = -24.62 + 0.25)

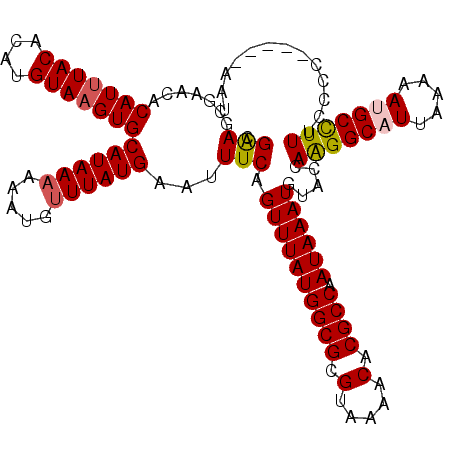
| Location | 13,313,999 – 13,314,114 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -25.41 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13313999 115 - 22224390 AUU-----GGGGAAAGGCAUUUUUAAUGCCCUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCG ...-----((.(((.(((((.....)))))....((((((((.(((((((.....)))))))............((((((.....))))))(((......)))))))))))))).))... ( -28.30) >DroVir_CAF1 64752 109 - 1 A-------AAGCAAAAGCAU-UUAAAUGCCUUGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUU--UUAUGCAC-UACAUGUGUAAAUGUGUCUCCUCCG .-------........((((-....))))...(((.(((((((.((((((.....))))))))))).))..........(((((--...(((((-.....))))))))))......))). ( -25.10) >DroPse_CAF1 74838 120 - 1 AGUAGCGAGGAGCAAGGCGUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGGUUCU ........(.(.((((((((.....)))))))).).)......(((((((.....)))))))..(((((((........(((((.....(((((......)))))))))).))))))).. ( -33.70) >DroEre_CAF1 18305 115 - 1 AUU-----GGGGAAAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCG ...-----((((.(((((((.....)))))))..((((((((.(((((((.....)))))))............((((((.....))))))(((......)))))))))))....)))). ( -30.50) >DroYak_CAF1 17770 115 - 1 AUU-----GGGGAAAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCG ...-----((((.(((((((.....)))))))..((((((((.(((((((.....)))))))............((((((.....))))))(((......)))))))))))....)))). ( -30.50) >DroPer_CAF1 74767 120 - 1 AGUAGCGAGGAGAAAGGCGUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGGUUCU .(((.(((((..((((....))))....))))).)))......(((((((.....)))))))..(((((((........(((((.....(((((......)))))))))).))))))).. ( -32.40) >consensus AUU_____GGGGAAAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCG .............(((((((.....)))))))..(((((((..(((((((.....)))))))............................((((......)))))))))))......... (-25.41 = -25.68 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:59 2006