
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,312,843 – 13,312,956 |
| Length | 113 |
| Max. P | 0.663770 |

| Location | 13,312,843 – 13,312,956 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -16.47 |
| Energy contribution | -17.94 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13312843 113 + 22224390 --CUGAACUUUGGCUCU-UGCCAAUUUUCUUUUGCCCAUUGUUUGGCUAUUUCUUUGGAUCGCUGAGCAACCUUUGACCUUAUCGACUCUCUUGG----UCGGGCGCAAUUAUCACCCCA --..(((..(((((...-.)))))..)))....((((.(((((..((.(((......))).))..))))).....((((..............))----))))))............... ( -32.84) >DroSec_CAF1 25637 110 + 1 --CUGAACUUUGGCUCU-UGCCAAUUUUCUUUUGCCCAUUGUUUGGCUAUUUCUUUGGACCGCUGAGCAACCUUUGACCUUAUCGACUCUC--GG----UCGGGCGCAAUUAUCACCCC- --..(((..(((((...-.)))))..)))....((((.(((((..((....((....))..))..))))).....((((............--))----))))))..............- ( -30.80) >DroSim_CAF1 17862 111 + 1 --CUGAACUUUGGCUCU-UGCCAAUUUUCUUUUGCCCAUUGUUUGGCUAUUUCUUUGGACCGCUGAGCAACCUUUGACCUUAUCGACUCUC--GG----UCGGGCGCAAUUAUCACCCCA --..(((..(((((...-.)))))..)))....((((.(((((..((....((....))..))..))))).....((((............--))----))))))............... ( -30.80) >DroEre_CAF1 17175 111 + 1 --CUGAACUUUGGCCCU-UGCCAAUUUUCUCUGGCCCAUUGUUUGGCUAUUUUUUUGGUCCGCUGAGCAACCUUUGACCUUAUCGACUCUU--GG----UCGGGCGCAAUUAUCACCCCA --..(((..(((((...-.)))))..)))..((((((.(((((..((..............))..))))).....((((............--))----)))))).))............ ( -30.14) >DroYak_CAF1 16589 113 + 1 CUCUGAACUUUGGCUCU-UGCCAAUUUUCUUUUGGCCAUUGUUUGGCUAUUUCUUUGGUGCGCUGAGCAACCUUUGACCUUAUCGACUCUG--GC----UUGGGCGCAAUUAUCACCCCA ....(((..(((((...-.)))))..)))...((((((.....))))))........((((.(..(((.....((((.....)))).....--))----)..)))))............. ( -31.90) >DroPer_CAF1 73795 108 + 1 --CUGAACUUUUCCACUUUUCCACUUUUCCUCUUUCUAUUGU--GGAAAUUCCC------AGCCAUAUCCCGUUUGACCUUAUCGACUUUU--GUAAGCCUCGGCACAAUUAUCGCCCCA --(((....(((((((........................))--)))))....)------)).....................(((...((--((..((....))))))...)))..... ( -13.86) >consensus __CUGAACUUUGGCUCU_UGCCAAUUUUCUUUUGCCCAUUGUUUGGCUAUUUCUUUGGACCGCUGAGCAACCUUUGACCUUAUCGACUCUC__GG____UCGGGCGCAAUUAUCACCCCA ....(((..(((((.....)))))..)))....((((.(((((((((..............)))))))))...((((.....))))...............))))............... (-16.47 = -17.94 + 1.47)

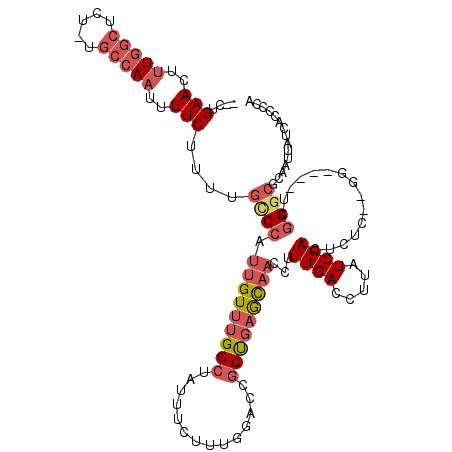
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:57 2006